1 Introduction
Using neutrons as probes to investigate biological entities has led to numerous significant breakthroughs in research fields at the interface between medicine, chemistry and physics [1–9]. The power of neutron investigation comes from the unique physical properties of these particles, which are used through a vast array of techniques to study different characteristics of matter in its various forms.
Instrumental Neutron Activation Analysis (INAA) is for example used to evaluate the concentration of trace and major elements [10–28] in materials. Toxic elements, such as As or Sb, can be detected in biological materials with a lower limit close to 0.005 μg/g [23]. The content of metals in various parts of the human brain has been discussed [24,27], through the assessment of the relationship between the content of Al and Alzheimer's disease [25].
Because of the neutron mass, the neutron wavelength is comparable to interatomic distances in matter, and neutron diffraction can be used to study crystal structures, that is, the arrangement of atoms in a crystalline material. Such information is of major importance in the case of implants made of bioceramics for bone filling and replacement [29–32]. More generally, because neutrons do not damage biological materials, they offer the opportunity to investigate bulk biological samples, whether as an aqueous solution, a solid, a powder, or a crystal. In addition, the neutron carries a spin, allowing one to study the static and dynamic magnetic properties of matter. As underlined by S.C. Vogel [33], neutron scattering uniquely complements other characterization techniques, and together with X-ray synchrotron, is considered amongst the most useful microscopic probes of matter available today.
The aim of this review is to give clinicians an overview on how neutron diffraction can be used to study different kinds of biological entities.
2 Neutron–matter interaction: some basic notions
Neutrons do not exist naturally in free form, but decay into protons, electrons, and anti-neutrinos. In order to produce neutrons, two kinds of sources exist and are based either on fission, in a conventional nuclear reactor, or on spallation (neutrons are obtained by bombarding a target of heavy elements with high-energy particles, typically, accelerated protons) processes.
2.1 Instrumental neutron activation analysis
When a sample is subjected to a high neutron flux, it emits gamma rays, following the decay of radioactive elements which have been produced. The energy and intensity of these gamma rays are directly related to the nature and concentration of the elements present in the sample. This technique has been used to investigate different kinds of biological entities, such as hair [13], liver [14], kidney stones [15,21], gallstones [16], cortical and trabecular bones [17,18,22], human prostate [19], human nails [20], human thumbnails [26] or urine [28].
Several investigations have also been dedicated to kidney stones [34,35]. C. Koeberl and P.M. Bayer [34] have for example determined the presence of rare-earth elements in brain tissues and in kidney stones. Rare-earth elements have an exogenous origin, being mainly incorporated into the human body via ingestion of food or by inhalation of dust particles. The data suggest that rare-earth elements are trapped in kidney stones, by a slow accumulation process. In [35], D. Gompertz et al. have compared biochemical indicators of renal dysfunction with liver and kidney cadmium levels measured by neutron activation analysis in a group of 37 cadmium smelters.
2.2 Neutron powder diffraction (NPD)
Scattering processes between neutrons and atoms are also used by scientists to study matter [36–40]. Neutrons are electrically neutral and interact only weakly with matter, and have, as a result, a penetration depth of several centimeters. Unlike X-rays, they do not interact with the electronic cloud of an atom, but with its nucleus: the strength and character of this neutron–nucleus interaction is characterised by the so-called neutron scattering length. This parameter varies irregularly from one nucleus to another. In particular, it is quite large for light elements like hydrogen, carbon, nitrogen or oxygen, which are almost invisible to X-rays (Fig. 1). Neutrons are thus an important tool in the investigation of the location and dynamics of protons, an essential aspect in many biological processes.

Neutron and X-ray scattering lengths for different atoms.
Elastic scattering, or diffraction, describes a process in which the incident and the scattered neutrons have the same energy. The scattering angle θ (as defined in Fig. 2) gives information, through Bragg’s law, on the periodicity of the atomic arrangement (i.e. crystal unit cell parameters). The intensity of the Bragg peaks is related to the cell symmetry and atomic positions within the crystal unit cell.

(a) Schematic representation of the Bragg law in diffraction, 2dhkl sin θ = λ, relating the diffraction angle θ, the beam wavelength λ, and the space between lattice planes (in thick dark lines) dhkl. (b) Rietveld refinement result of the NPD diagram of a cystine kidney stone (experimental data Yobs in red, calculated profile Ycalc in black, and Yobs−Ycalc difference in blue). Tick marks in green below the profiles indicate the peak positions of allowed Bragg reflections for cystine.
As shown in Fig. 2, a neutron powder diffraction (NPD) pattern is simply a recording of the intensity of diffraction (in neutron counts) versus the diffraction angle θ.
A typical example of a neutron powder diffractometer is the G4.1 instrument, located at the French neutron source Orphée. The Orphée reactor produces a white neutron beam (broad distribution of wavelengths) and a monochromator is used to select the desired wavelength. The monochromator is commonly a single crystal of germanium or silicon; on the G4.1 beamline, a vertically focusing pyrolytic graphite monochromator is used. The detector is an 800-cell multidetector covering a 2θ range spanning 80°, which collects all reflections simultaneously, thus increasing the data acquisition speed (Fig. 3).
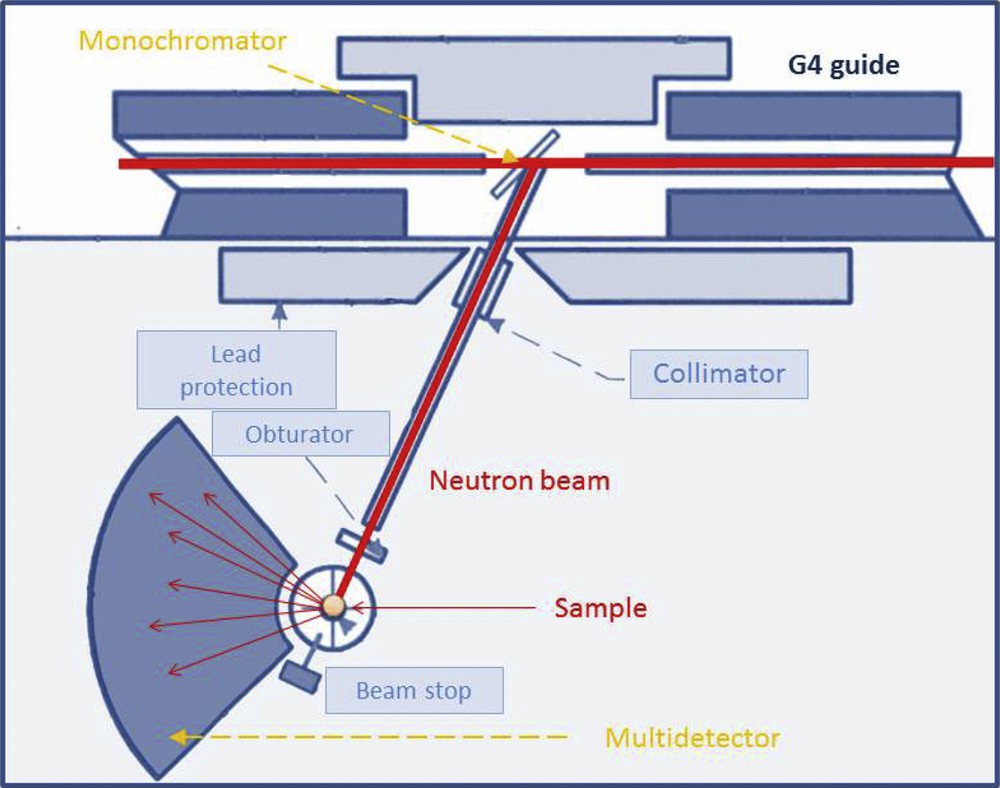
Schematic representation of the G4.1 beamline.
Rietveld refinement [41] is the most common method used for the determination of crystal structures from X-ray or neutron diffraction data. A theoretical diffraction pattern is calculated based on a proposed model, and compared with the experimental data. Gradual changes are introduced into the model, to minimize the difference between the observed and calculated patterns (Fig. 2). This is usually performed using a computer software package such as FullProf, Jana2006, or GSAS [41].
Rather straightforward experimentally, powder diffraction provides after data refinement a wealth of information, not only about the detailed atomic arrangement, but also about the size, the residual strains and the morphology of the powder particles. For instance, the broadening of the diffraction peaks can be the result of two physical factors, either a small particle size or residual microstrain, whose effects can be analyzed separately [42]. The size of nanocrystals can therefore be estimated after analysis of the peak profiles. This can be of clinical importance, as in [43], in which U. Vetter et al. have pointed out significant modifications of the apatite crystal sizes in the bones of patients with osteogenesis imperfecta.
Regarding the orientation of the nanocrystals within a bulk material, diffraction also provides key information: Bacon and Goodship [44] have shown through neutron diffraction that the long bones of animals have the c-axes of their apatite crystals preferentially oriented to withstand stresses. Recent experimental developments combining a monochromatic X-ray beam (<30 μm) with a charge-coupled device camera allow a relatively easy quantification of crystal alignment in bone tissue [45].
3 Neutron scattering and kidney stones
Biological calcified entities such as kidney stones show several levels of organization. They result from an agglomeration of crystallites of a few micrometers, each crystallite being made of nanocrystals (whose dimensions are about a few hundreds of nanometers, and can be studied by neutron scattering). The definitions for the terms ‘nanocrystals’ and ‘crystallites’ are made according to Van Meerssche & Feneau-Dupont [46].
3.1 Cystine kidney stones
Recently we have published a neutron powder diffraction (NPD) and FE-SEM investigation on cystinuria [47]. Worldwide, the incidence of cystinuria is about 1:7000 births and represents a major medical problem, because of stone recurrence and the risk to induce renal failure. This genetic disease comes from a mutation in renal epithelial cell transporters [48–53]. More precisely, two different types of cystinuria may be distinguished according to genetics: type A cystinuria, due to mutations along the SLC3A1 gene encoding the rBAT heavy subunit, and type B cystinuria, with mutations in the SLC7A9 gene encoding for light subunit b0,+ AT [54]. In both A and B cystinuria, mutations result in a deficient reabsorption of the dibasic amino acids and cystine [54,55]. The excessive amount of cystine in urine leads to a high risk of cystine precipitation, and to the formation of stones as a consequence of the poor solubility of this amino acid.
Regarding cystine kidney stones, two morphological types, namely Va and Vb, exist [56–58]. The surface of the first kind of stone (type Va, Fig. 4a) is homogeneous, granular, made of crystals with blunt angles, and is commonly observed in untreated cystinuric patients. When cystinuria is treated with alkali or/and thiol therapy, a second type of cystine kidney stone may appear (type Vb, Fig. 4b).

(a) Va cystine kidney stone, (b) Vb cystine kidney stones. The cross-section of a stone reveals a diffuse concentric structure at the periphery, and an unorganized agglomerate of cystine crystals in the core, (c) typical neutron diffraction diagrams of cystine kidney stones collected on the G4.1 beamline. (d) Broadening of a Bragg peak, corresponding to the decrease of the crystallite size after treatment with sodium carbonate alkaline therapy, which evolves from >200 nm (Va) to ∼138 nm (Vb) [58].
The salient point of this study comes from NPD, and lies in the result that only the usual alkaline therapy with sodium bicarbonate reduces significantly the growing process of cystine crystals. All the other therapies, namely alkalinization with other salts (Foncitril, Alcaphor, potassium citrate) and thiol therapy (sulfhydryls) based on tiopronine, d-penicillamine or on captopril treatments, seem inefficient. A similar conclusion is obtained through FE-SEM observations. In conclusion, all observations agree that only the usual alkaline therapy with sodium bicarbonate reduces the size of both crystals and crystallites, while other medical treatments only alter the surface of the crystallites.
3.2 Calcium oxalate kidney stones
In western countries, calcium oxalate (CaOx) is the main component of more than 70% of all kidney stones [59,60]. Among the different crystalline forms, calcium oxalate monohydrate (COM – CaC2O4–H2O), or whewellite, is the most frequent [61–63]. COM kidney stones are related to hyperoxaluria (too much oxalate present in the urine), while excess of calcium (hypercalciuria) in urine leads to the formation of calcium oxalate dihydrate (COD – CaC2O4–2H2O) [64]. Of note, a third species exists, namely calcium oxalate trihydrate (COT – CaC2O4–3H2O – caoxite), which has been observed – rarely – in patients treated with drug containing a precursor of oxalate [65].
Hyperoxaluria may have different origins, from dietary to genetic [57]. At the macroscopic scale, significant differences exist. Intermittent hyperoxaluria of dietary origin corresponding to idiopathic stone formers leads to stones with a dark-brown smooth or mammillary surface; for primary hyperoxaluria [66–72] stones have a surface with a mulberry-shaped aspect of cream or pale yellow–brown color [57]. These different kinds of hyperoxaluria lead to the same whewellite chemical phase.
To compare the structural characteristics of the stones at the mesoscopic and nanometric scale, NPD and FE-SEM studies were carried out [73]. The crystal structure of whewellite was first revisited using single-crystal neutron diffraction at room temperature. Then, whewellite stones corresponding to different kinds of hyperoxaluria were examined, at the mesoscopic scale with FE-SEM, and at the nanometric scale with NPD (Fig. 5). The results show that, although the various whewellite kidney stones exhibit similar neutron diffraction patterns, significant differences exist at the mesoscopic scale, which explains the polymorphism of whewellite stones observed at the macroscopic scale. Combined NPD and FE-SEM datasets therefore define a completely new medical diagnosis of primary hyperoxaluria [74], a rare inherited disease, leading to recurrent nephrolithiasis, nephrocalcinosis, systemic oxalosis, and renal failure, ultimately requiring combined kidney and liver transplantation.
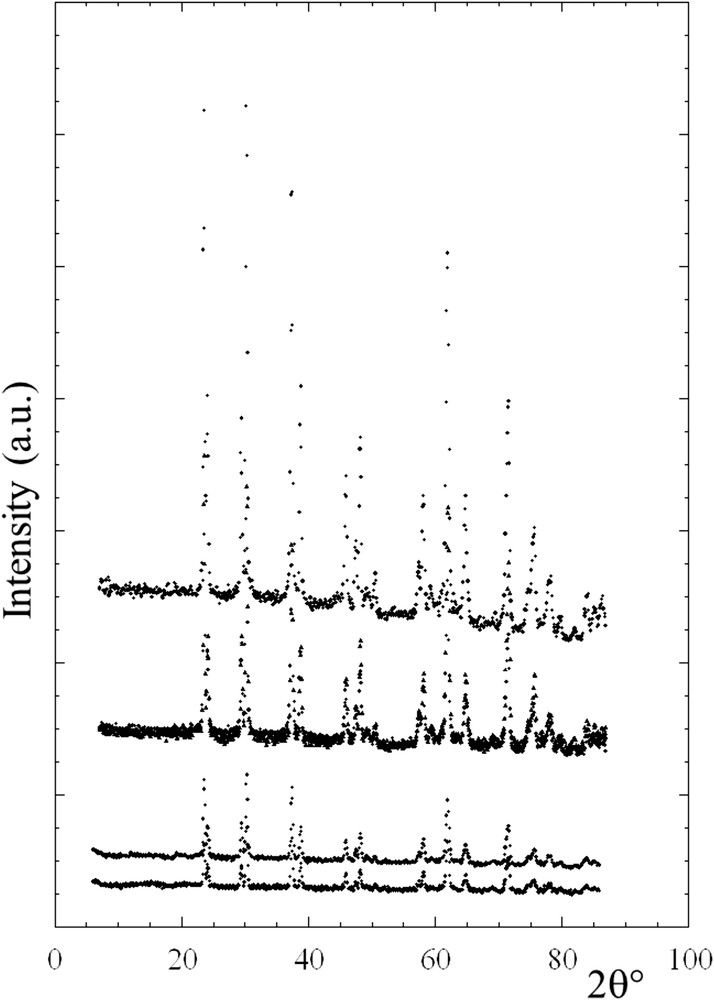
Different scattering diagrams collected on G4.1 on whewellite kidney stones.
3.3 Struvite kidney stones
Unlike more common calcium kidney stones, struvite stones [75–80] are formed by bacterial waste products during a kidney or urinary tract infection (UTI). Struvite kidney stones can lead to rapid obstruction, hydronephrosis, recurrent pyelonephritis, and decreased kidney function; owing to the urine decomposition process, large (cm) kidney stones can grow over a period of a few weeks. Urea-splitting bacteria such as Proteus, Staphylococcus, Pseudomonas, Providencia, and Klebsiella generate struvite kidney stones [81–83]. In addition, urease-negative bacteria like those with weak urease activity, such as some strains of Escherichia coli, may also be involved in the formation of struvite renal calculi, although urease-positive bacteria seem to play a greater role. If these organisms leave their imprints at the surface of kidney stones made of apatite, the presence of such imprints has not been found at the surface of struvite kidney stones. Moreover, in a stone containing both carbonated apatite and struvite components, bacterial imprints were observed on the carbonated apatite but not on the struvite part.
NPD can provide an understanding of this apparent contradiction between clinical data gathered at the hospital and observations obtained through FE-SEM [84]. A typical diffraction pattern collected for a struvite kidney stone is compared in Fig. 6 with the pattern of an apatite kidney stone. Even if the background level was high (owing to the large hydrogen content of the biological samples), the high quality of the signal-to-noise ratio allows one to perform a complete Rietveld-type refinement, with an analysis of the crystallite size for both samples. While struvite kidney stones consisted of quite large nanocrystals (250 nm), as expected from the well-defined diffraction peaks (Fig. 6), crystallites in carbonated calcium phosphate apatite kidney stones are much smaller, ∼50 nm. To explain the absence of bacterial imprints at the surface of struvite kidney stones, an analogy with a man walking on a beach can be drawn: if the beach is made of sand, footprints may be observed, but if the beach is made of stone, no footprints will be seen. Therefore, a relationship between the size of bacteria and the size of nanocrystals becomes relevant when looking for bacterial imprints: bacterial imprints may appear with small calcium carbonated apatite nanocrystals, rather than with large struvite nanocrystals. This result obtained through neutron diffraction has a major clinical significance. Neutron diffraction diagrams validate an approach in which infection leads to the presence of struvite kidney stones or the presence of bacterial imprints at the surface of Ca phosphate apatite even if infection is asymptomatic. Indeed, for several patients without any symptoms related to infection (negative urinary test, no fever…) and which have kidney stones without struvite or Ca apatite with a high carbonation rate, observations through Field Emission Scanning Electron Microscopy (FE-SEM) point out the presence of bacterial imprints. Antibiotic therapy starts immediately, avoiding major kidney infection, which can lead to the loss of the kidney function and thus to dialysis and kidney graft.

Neutron diffractograms collected on G4.1 for struvite and calcium phosphate apatite kidney stones.
3.4 Apatite kidney stones and ab initio simulation
Regarding nanometer scale particles, it is also possible to build neutron (or X-ray) diffraction intensity I(q) through Debye scattering [85–91].
In this equation, I(q) is the angle dependent intensity from coherent scattering, the sums over i and j are over all the atoms, Rij is the distance between the atoms i and j, and bi and bj are the neutron scattering lengths of the different atoms i and j. It is quite easy to build a structural model for particles displaying different morphologies (cylinders, spheres, etc.) and to calculate the scattering intensity. More precisely, A. Guagliardi et al. [92] have used the hexagonal P63/m structure [93–95] as the building block for constructing a family of nanocrystals.
Following this approach, it is possible to calculate scattering profiles for different particle shapes, and match them to experimental data (Fig. 7, in the case of nanocrystals of Ca phosphate apatite). The next step will be to study the effect of other parameters, such as cationic substitution in the nanocrystals investigated.

Scattering diagrams calculated following the Debye formulae for different morphologies (cylinders and spherical) compared with experimental data.
4 Neutron scattering and bioceramics
Ceramics such as Al2O3/Y–ZrO2 [96] and Ca phosphates [97–102], as well as bioactive glasses [103–105], are extensively used in different kinds of prosthesis. Their behaviour during manufacturing and in the human body is therefore of great importance and a topic of extensive research. Being able to understand and predict their behaviour under stress or strain is a major challenge, which can be achieved with neutron diffraction.
Regarding Ca phosphate, several investigations have been performed on the Ca phosphate apatite using the neutron probe. For example, based on inelastic neutron-scattering experiments, C.K. Loong et al. [106] underlined the lack of OH− ions in the crystals of bone apatite. R.G. Hill et al. [107] have investigated the early stages of nucleation and crystallisation of an apatite (fluorapatite, Ca5(PO4)3F). P.C.H. Mitchell et al. [108] have assessed the localisation of water in the apatite framework. More recently, D. Arcos et al. [109,110] have investigated silicon doped hydroxyapatites (SiHA) by neutron diffraction. They have noticed an enhanced disorder of the H atom belonging to the OH groups in SiHA, which could contribute to the higher reactivity of SiHA with respect to HA. Finally, combining neutron scattering experiments and NMR, S. Gomes et al. [111] have underlined the presence of two types of protons in the Si-substituted HAp phase, the new site corresponding to species engaged in hydrogen bonding with silicate anions.
5 Conclusion and perspectives
Using neutrons as probes allows clinicians to gather information on trace elements [112], and to get structural information, both at the atomic scale (crystal structure) and at the crystallite scale (crystallite size). The fact that this technique is non-destructive, and can investigate bulk elements while preserving the physicochemical state of the biological samples, constitutes a unique and precious advantage. With the micro-beam instruments available at the new European neutron source [113] and progress in neutron tomography, mapping “bulk” biological samples will also allow new types of investigations, like the structural characteristics of nanopathological calcifications embedded in tissues, for example. Neutron spectroscopy is another technique that can be evoked, as it is already used extensively in biology, to study, e.g., water dynamics in cells, or protein unfolding.


