1. Introduction
Population growth and rapid industrialization have led to severe problems of environmental contamination, in particular by heavy metals [1, 2]. These chemicals pose threats to human health and ecosystem due to their mobility, toxicity, persistence and bioaccumulation [3].
Chromium is a toxic heavy metal derived from both natural and anthropogenic sources. This metal has recently received widespread attention because of its interesting proprieties. It is used in many industrial and defense applications such as stainless steel, electroplating, leather tanning, textile dyeing, dyes, pigments, nuclear weapons production, and metal processing industries [4, 5]. Chromium exists in a wide range of valence states from Cr(II) to Cr(VI) [6], but in nature it generally exists in two stable oxidation states, trivalent chromium Cr(III) and hexavalent chromium Cr(VI) [6]. The Cr(VI) compounds are 100-fold more toxic than Cr(III) compounds due to their high solubility in water, rapid permeability through biological membranes and subsequent interaction with intracellular proteins and nucleic acids [4]. Hexavalent chromium has been classified as one of the priority pollutants, that poses the greatest threat to humans, by several regulatory agencies including the United States Environmental Protection Agency (USEPA) [7]. Besides, the World Health Organization has fixed the maximum concentration limits in drinking water of total chromium and hexavalent chromium at 2.00 and 0.05 mg⋅L−1 respectively [8]. At high concentrations, Cr(VI) could cause environmental and health problems because of its high toxicity [6]. Chronic exposure to Cr(VI) in humans may cause hemorrhage, allergies, respiratory tract disorders, ulceration, epigastric pain, cancer in the digestive tract and lungs and birth deficiency [1]. Even at low concentrations, this heavy metal may produce mutagenesis and carcinogenic diseases [1, 9]. Cr(VI) could be also harmful to flora and fauna of natural aquatic ecosystems. Indeed, previous studies [9] have shown that short-term Cr(VI)-exposure could inhibit some biological activities of the soil and decrease the microbial biomass whereas long-term exposure could negatively affect the soil microbial community, reducing the microbial biomass as well as its activity and diversity.
Several conventional procedures have been established for the removal of chromium from polluted environments [10]. Traditionally, these techniques reduce Cr(VI) concentrations to levels that comply with standards or convert Cr(VI) to Cr(III) and were applied in both in situ and ex situ systems [6]. Some of the physico-chemical processes used include precipitation, ultrafiltration, membrane separation, electrocoagulation, solvent extraction, ion exchange, reverse osmosis and adsorption [11]. However, the currently used methods are often complicated and present disadvantages due to their tendency to cause secondary pollution, long processing time and high costs of equipment, reagent, and energy requirements [11]. Therefore, there has been a trend toward researching a better alternative, a low cost, and eco-friendly method for treatment of organic and inorganic pollution [12, 13, 14, 15].
The development of suitable methods for the remediation of Cr(VI)-contamination is nowadays considered as an important topic [16]. The use of passive biological processes such as bacterial biomass could ensure recovery and long-term protection of the environment [6]. In this regard, bioremediation through microorganisms is getting importance day by day and this has been so ever since the discovery of the first microbe capable of reducing Cr6+ in the 1970s [17]. The direct use of bacterial biomass with distinctive features is a novel, efficient, eco-friendly, and cost-effective approach for remediation of heavy metals in general and chromium in particular [18, 19, 20]. The in situ biotransformation of Cr(VI) to Cr(III) for effective removal of Cr(VI) from water has been demonstrated in both laboratory and pilot systems but has not been applied on the full scale [21]. The design and operation of biotechnological processes for Cr(VI) reduction involve a thorough understanding of the effect of Cr(VI) on the kinetic characteristics of the Cr(VI) reduction processes and the evaluation of different parameters affecting the effectiveness and profitability of these processes [22, 23].
Many factors can influence the reduction of chromium. Cheng and Li [5] investigated the influence of reaction temperature, pH, and Cr(VI) concentration and found that Cr(VI) reduction was optimum at 37 °C and pH 8. Liu et al. [24] reported that the reduction of Cr(VI) is greatly affected by the used pH and Cr(VI) initial concentration. The maximum reduction rate was observed at pH 9 under a wide range of concentrations from 10–80 mg⋅L−1.
Caravelli and Zaritzky [25] studied the effect of chromium initial concentration on the biological activity of the S. natans biomass, specifically on its reductase activity and found that high concentrations of Cr(VI) and biomass significantly enhanced the Cr(VI) reduction process. Also, Rath et al. [26] investigated the influence of initial concentration of Cr(VI) using bacterium Bacillus amyloliquefaciens (CSB 9) and observed an inhibitory effect of higher concentration on Cr(VI) reduction. All these studies were conducted using classical one-variable-at-a-time experimentation. These reports indicate that the reduction of chrome is mainly affected by temperature, pH and initial Cr concentration. Additional factors such as biomass density, carbon source, dissolved oxygen, oxidation–reduction potential, presence of other oxyanions, and other metal cations and their combined effects could play a major role for a better understanding and controlling of the involved biotechnological processes in Cr(VI) reduction [27]. Studying each operating parameter separately to investigate how it affects the bioremediation process will require considerable time, chemicals and human energy. Furthermore, the interactive effects of dominant parameters will not be discernible when applying the traditional one-factor-at-a-time method. Hence, it is important to use an effective method that reduces significantly the number of experiments with time-saving, energy, cost and which is able to assess both single and interactive effects of process variables. In this regard, statistical experimental design is an appropriate approach for evaluating accurately the most important operating parameters and their interactions on the process efficiency [28]. It enables much reduction in terms of time, effort and chemicals with a limited number of planned experiments [29]. The use of full factorial designs can improve the bioremediation effectiveness. In fact, this approach generates maximum information about the factors, defining the relevant ones and identifying the interactions among them. Also, it predicts the effect that such interactions could have on the experimental response [30].
For that purpose, the present research was developed in order to: (i) isolate and identify indigenous chromium-resistant microorganisms from polluted soils and sludge, (ii) optimize and investigate the effect of the crucial environmental parameters such as temperature, Cr(VI) initial concentration, pH and time on chromium removal using a full factorial design at two levels. A mathematical model was established allowing fixing experimental conditions for each desired degradation of chrome by the selected bacterial strain.
2. Materials and methods
2.1. Isolation of Cr-resistant bacteria
Chromium-resistant bacterial strains were isolated from polluted soils and sludge in an industrial zone located in the Sidi Bel Abbes city (35° 11′ 38′′ N, 0° 38′ 29′′ W), Algeria.
Fifteen soil and sludge samples, weighing about 30 g each, were collected in sterile bags from two sampling sites: the first one from near the National Company of Electronic Industries (ENIE), the second from near the production site of Manufacturing and Sale of Agricultural Equipment (SONACOM).
All chemicals used were of analytical grade. Nutrient broth and agar plates used for bacterial growth enrichment and isolation were prepared using sodium chloride (NaCl 5 g⋅L−1), Peptone A (5 g⋅L−1), beef extract (3 g⋅L−1), and agar (1.5 g) for 100 mL of the nutrient broth medium. The pH of the bacterial growth media was maintained at 6.8. All media were autoclaved at 121 °C and 1 bar pressure for 15 min [12, 31]. 1 mM K2Cr2O7 as Cr(V) source, was added to the culture medium. The growth of the bacterial colonies was observed after 24 h of incubation at 30 °C under aerobic conditions.
2.2. Screening of resistant isolates for Cr reduction (biomass quantification)
To select the most effective strains, screening was carried out for Cr-resistant bacteria and their chromium removal ability. All the obtained isolates were screened at first for tolerance against Cr(VI) and the most promising microorganisms (those which gave maximum bacterial biomass) were selected to be screened for chromium(VI) reduction. For conducting the screening experiments under aerobic conditions, shake flasks with cotton plugs were used. Flasks with 100 mL of culture medium amended with Cr(VI) solution, were inoculated with 1 mL of bacterial suspensions prepared previously for each isolate in sterile nutrient broth. The optical density (OD) of all the bacterial suspensions was adjusted at 0.5 McFarland at 600 nm to save the same conditions of initial biomass (106 cell∕mL). Un-inoculated flasks containing only the nutrient broth and Cr(VI) served as control. The experiments were carried out at 30 °C, 120 rpm for 24 h.
2.3. Evaluation of Cr(VI) reduction potential of the bacterial isolates
The reduction potential of the selected chromate-resistant strains was evaluated under aerobic conditions at 30 °C and 120 rpm with an initial Cr(VI) concentration of 1 mM in a sterile nutrient broth. The bacterial cell density of the liquid cultures was routinely monitored by measuring the OD at 600 nm. Cr(VI) concentrations were determined by the 1-5 diphenylcarbazide (DPC) method using a spectrophotometer (OPTIZEN POP UV/Vis) at 540 nm [32].
2.4. Identification of Cr(VI)-removing bacteria
The most effective strain for chromium removal was identified by analyzing their total proteins (ribosomal proteins and proteins associated with membranes) using the MALDI-TOF/MS BIOTYPER from Bruker Daltonic (Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight) [33]. This method analyses the displacement of ionic entities in electromagnetic fields. A mixture of matrix-sample co-crystallized on a metal surface or target is subjected to the firing of a laser beam (337 nm, 20 Hz) to its desorption and ionization. The time of flight of the generated ions is measured and this allows obtaining a mass spectrum [34].
2.5. Experimental design and statistical analysis
The statistical software Design-Expert® (version 7.0.0, Stat-Ease, Inc.) was applied in the present study to estimate the importance and the interaction levels of four operational variables: temperature, initial pH, chromium concentration and contact time. A 24 full factorial design [35] was performed to design all possible experiments, analysis of data and interpretation.
The significance of the main effects and their interactions on the responses were evaluated with ANOVA where the P values were generated to prove the null hypothesis with 95% confidence level. A first-order polynomial regression equation was chosen to fit the experimental results (1).
Through the applied factorial model, each parameter represented in (1) can be determined from a limited number of experimental runs [35]
| (1) |
Factor levels in the 24 factorial experimental design
| Factor | Unit | Symbol | Levels | |
|---|---|---|---|---|
| −1 (Low) | +1 (High) | |||
| Temperature | °C | A | 25 | 55 |
| Concentration of Cr | mM | B | 0.5 | 1.5 |
| pH | / | C | 3 | 9 |
| Time | H | D | 20 | 180 |
The list of investigated independent variables with their coded and actual levels are presented in Table 1. Low and high levels of each variable were coded as: −1 and +1.
The 3D surface plots were generated using the applied software in order to explain relationships between responses and each pair of the four independent variables for achieving the highest response.
Screening of the most Cr-resistant strains (biomass quantification)
| B.S.* | Biomass quantification of bacterial strains** | |||
|---|---|---|---|---|
| 0 h | 12 h | 24 h | ||
| S1 | C1 | 0 | 0.201 | 0.023 |
| C2 | 0 | 0.02 | 0.002 | |
| C3 | 0 | 0.011 | 0.023 | |
| C4 | 0 | 0.012 | 0.006 | |
| S2 | C1 | 0 | 0.023 | 0.022 |
| C2 | 0 | 0.01 | 0.003 | |
| S3 | C1 | 0 | 0.002 | 0.006 |
| C2 | 0 | 0.071 | 0.073 | |
| C3 | 0 | 0.006 | 0.01 | |
| C4 | 0 | 0.011 | 0.059 | |
| S4 | C3 | 0 | 0.124 | 0.053 |
| S5 | C1 | 0 | 0.023 | 0.056 |
| C2 | 0 | 0.091 | 0.09 | |
| C3 | 0 | 0.031 | 0.015 | |
| C4 | 0 | 0.013 | 0.005 | |
| S6 | C2 | 0 | 0.056 | 0.07 |
| C4 | 0 | 0.006 | 0.007 | |
| C5 | 0 | 0.004 | 0.008 | |
| S7 | C2 | 0 | 0.017 | 0.014 |
| C4 | 0 | 0.009 | 0.002 | |
| S8 | C1 | 0 | 0.007 | 0.001 |
| C3 | 0 | 0.014 | 0.004 | |
| C4 | 0 | 0.005 | 0.002 | |
| C5 | 0 | 0.002 | 0.01 | |
| S9 | C1 | 0 | 0.017 | 0.002 |
| C2 | 0 | 0.002 | 0.008 | |
| C3 | 0 | 0.008 | 0.008 | |
| C5 | 0 | 0.009 | 0.051 | |
| C6 | 0 | 0.01 | 0.009 | |
| C7 | 0 | 0.005 | 0.007 | |
| S10 | C1 | 0 | 0.012 | 0.104 |
| C2 | 0 | 0.005 | 0.037 | |
| C3 | 0 | 0.002 | 0.002 | |
| C4 | 0 | 0.002 | 0.002 | |
| C6 | 0 | 0.024 | 0.027 | |
| S11 | C3 | 0 | 0.001 | 0.006 |
| C5 | 0 | 0.048 | 0.05 | |
| S12 | C1 | 0 | 0.005 | 0.004 |
| C2 | 0 | 0.039 | 0.04 | |
| C3 | 0 | 0.016 | 0.026 | |
| C4 | 0 | 0.002 | 0.003 | |
| C5 | 0 | 0.008 | 0.002 | |
| C6 | 0 | 0.036 | 0.053 | |
| S13 | C1 | 0 | 0.008 | 0.103 |
| C2 | 0 | 0.084 | 0.043 | |
| C3 | 0 | 0.076 | 0.093 | |
| S14 | C1 | 0 | 0.006 | 0.01 |
| S15 | C1 | 0 | 0.07 | 0.072 |
| C2 | 0 | 0.0 | 0.003 | |
| C3 | 0 | 050.079 | 0.078 | |
| C4 | 0 | 0.049 | 0.044 | |
| C5 | 0 | 0.02 | 0.041 | |
| C6 | 0 | 0.045 | 0.05 | |
| C7 | 0 | 0.014 | 0.039 | |
*Bacterial strain, **optical density measure.
3. Results and discussions
3.1. Isolation and screening of Cr-resistant strains for chromium (VI) reduction
A total of 54 chromium resistant bacterial strains were isolated from 15 industrial locations (contaminated soils and sludge samples) in nutrient broth medium amended with 1 mM of K2Cr2O7 as Cr(VI). The distribution pattern of the Cr-resistant strains is given in Table 2. Four of these 54 isolated strains showed resistance to Cr toxicity with high production of bacterial biomass. This behavior confirmed that some bacteria develop a stress response and an auto defense mechanism against heavy metals inducing the formation of heavy metal resistant bacteria, able to survive in the adverse conditions [36, 37].
3.2. Cr reduction ability of the Cr-resistant isolate strains
The four Cr-resistant isolate strains designed as S5C2, S10C1, S13C1 and S13C3 were selected to test their Cr(VI) reduction ability. Table 3 shows the corresponding chromium removal efficiency results. The chromium effect on bacterial growth and chromium reduction assays were also done in nutrient broth medium amended with 1 mM of K2Cr2O7 under aerobic conditions. The chromium removal rate by S10C1 was 61.5% after 12 days of incubation. Removal efficiencies observed with S13C1 and S13C3 were almost similar, both of the strains having removed about 40% of chromium. However, the performance of S5C2 was less than the rest of the strains with a removal efficiency of only 17.5%.
It is important to consider the fact that the microbial ability to reduce Cr(VI) depends on the medium composition and cell density [38]. Control tests without bacterial cultures showed that there was no significant reduction of Cr(VI) by abiotic processes with an average percentage reduction of 1.5% after 12 days. These results indicate that all tested strains were capable of reducing hexavalent chromium from the culture medium but the ability of S10C1 to remove Cr was remarkably higher as shown in Table 3. This strain was identified and used for the statistical analysis.
Previous studies have proved that microorganisms isolated from the contaminated sites showed high resistance toward pollutants and hence better pollution remediation potential [39, 40]. In fact, for an effective bioremediation approach it is important to have bacterial strains that combine a high resistance property and ability to reduce Cr(VI) [7]. In this regard, Garavaglia et al. [41] reported that Klebsiella oxytoca showed high tolerance and reduction potential of Cr(VI). Shakoori et al. [42] also indicated that Bacillus pumilus, Alcaligenes faecalis and Staphylococcus sp which were isolated from chromium-containing waste showed high Cr(VI) reduction capability. The same trend was observed for, Bacillus sp. JDM-2-1 and S. capitis [12], Bacillus sp. MDS05 [5], Acinetobacter sp. B9 [43], A. fumigatus and A. flavus [44], Brucella sp. [45], Hypocrea tawa [22], B. subtilis and B. safensis [46]. According to Das et al. [47], bacteria isolated from chromite mine soils have resistance to Cr(VI) and also to other heavy metals. All these results, in concordance with ours, confirmed that bacteria present in heavy metal contaminated soil exhibit remarkable resistance to metals and a high potential for reduction.
3.3. Identification of the most efficient strain
The isolate S10C1 was chosen because of its highest efficiency in removing Cr(VI) compared to the rest of the used strains. The test of gram-staining showed that it is Gram-positive. Coleman [48] reported that Gram-positive chromium reducing bacteria have significant tolerance to high concentrations of Cr(VI). The diameter of the colonies was 3–6 mm and has asymmetrical and crenate borders with matt surfaces. The identification of S10C1 strain by their molecular imprints was conducted using the MALDI-TOF/MS BIOTYPER. The analysis was performed by smearing a small amount of the organism from a single colony directly onto a spot on the MALDI-TOF MS steel anchor plate. This analysis was performed in automatic mode.
The results obtained from Figure 1 were analyzed on the NCBI taxonomy database (National Centre for Biotechnology Information) using the BLAST program (Basic Local Alignment Search Tool) to detect regions of similarity and compare nucleotide or protein sequences to sequence databases. So, S10C1 was identified as Bacillus cereus 4080 LBK (NCBI:txid1396) with a score value (based on pattern matching) of 1.927 in agreement with results published by TeKippe et al. [49] in which acceptable score was defined as ⩾1.7 for genus- and species-level identification of Gram-positive bacteria.
Representative mass spectrum of S10C1 bacterial strain.
Percentage reduction of Cr(VI) by the selected isolate strains according to the cultivation time
| Isolates | % chrome(VI) reduction | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cultivation time (day) | |||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| Control | 0 | 0 | 1.48 | 2.96 | 2.96 | 0 | 1.48 | 1.48 | 0 | 5.92 | 3.70 | 1.48 | 1.48 |
| S5C2 | 0 | 14.81 | 23.70 | 13.33 | 12.59 | 9.62 | 17.77 | 17.77 | 31.85 | 17.03 | 17.70 | 17.50 | 17.50 |
| S10C1 | 0 | 14.81 | 16.29 | 22.22 | 20 | 22.22 | 31.85 | 33.33 | 33.33 | 36.29 | 38.51 | 51.85 | 61.48 |
| S13C1 | 0 | 14.07 | 22.22 | 25.18 | 24.44 | 34.07 | 34.81 | 34.81 | 37.77 | 30.37 | 33.33 | 49.62 | 40.74 |
| S13C3 | 0 | 19.25 | 23.70 | 28.88 | 33.33 | 35.55 | 38.51 | 41.48 | 39.25 | 32.59 | 33.33 | 59.25 | 40.74 |
3.4. Experimental design and statistical analysis
3.4.1. Analysis of data, main effects of factors and model validity testing
A total of 16 experiments were carried out under the given experimental conditions and the values of the response Y (% removal) are illustrated in Table 4.
Design matrix for 24 full factorial design and corresponding responses
| Run | A | B | C | D | Responses* | ||||
|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | [Cr(VI)] (mM) | pH | Time (h) | Y (%) | |||||
| Coded | Actual | Coded | Actual | Coded | Actual | Coded | Actual | ||
| 1 | −1 | 25 | −1 | 0.5 | −1 | 3 | −1 | 20 | 59.6 |
| 2 | 1 | 55 | −1 | 0.5 | −1 | 3 | −1 | 20 | 92.9 |
| 3 | −1 | 25 | 1 | 1.5 | −1 | 3 | −1 | 20 | 30.0 |
| 4 | 1 | 55 | 1 | 1.5 | −1 | 3 | −1 | 20 | 52.7 |
| 5 | −1 | 25 | −1 | 0.5 | 1 | 9 | −1 | 20 | 4.4 |
| 6 | 1 | 55 | −1 | 0.5 | 1 | 9 | −1 | 20 | 18.4 |
| 7 | −1 | 25 | 1 | 1.5 | 1 | 9 | −1 | 20 | 7.4 |
| 8 | 1 | 55 | 1 | 1.5 | 1 | 9 | −1 | 20 | 9.9 |
| 9 | −1 | 25 | −1 | 0.5 | −1 | 3 | 1 | 180 | 98.9 |
| 10 | 1 | 55 | −1 | 0.5 | −1 | 3 | 1 | 180 | 99.5 |
| 11 | −1 | 25 | 1 | 1.5 | −1 | 3 | 1 | 180 | 57.6 |
| 12 | 1 | 55 | 1 | 1.5 | −1 | 3 | 1 | 180 | 79.3 |
| 13 | −1 | 25 | −1 | 0.5 | 1 | 9 | 1 | 180 | 39 |
| 14 | 1 | 55 | −1 | 0.5 | 1 | 9 | 1 | 180 | 19.1 |
| 15 | −1 | 25 | 1 | 1.5 | 1 | 9 | 1 | 180 | 20.7 |
| 16 | 1 | 55 | 1 | 1.5 | 1 | 9 | 1 | 180 | 1 |
*Percentage of chromium removal.
The determination of the significant main and interaction effects of factors affecting the Cr(VI) uptake capacity was followed by performing ANOVA on the two level factorial design. The results of ANOVA tests of the adopted regression model are presented in Tables 5 and 6. Fisher’s F-test, the distribution of the ratio between the respective mean square effect and the mean square error was used to evaluate the presence of a significant difference in relation to the control response and to calculate the standard errors. The p-values were used to identify experimental parameters that have a statistically significant influence on the specific response.
ANOVA results for chromium biodegradation
| Source | Sum of squares | DF* | Mean square | F-value | p-value prob > F | |
|---|---|---|---|---|---|---|
| Model | 17628.805 | 7 | 2518.400 | 36.000 | <0.0001 | Significant |
| A | 189.813 | 1 | 189.813 | 2.713 | −0.1381 | |
| B | 1873.310 | 1 | 1873.309 | 26.779 | −0.0008 | |
| C | 12699.769 | 1 | 12699.769 | 181.541 | <0.0001 | |
| D | 1223.758 | 1 | 1223.758 | 17.493 | −0.0031 | |
| AC | 642.052 | 1 | 642.052 | 9.178 | −0.0163 | |
| AD | 502.847 | 1 | 502.847 | 7.188 | −0.0279 | |
| BC | 497.256 | 1 | 497.256 | 7.108 | −0.0285 | |
| Residual | 559.643 | 8 | 69.955 | |||
| Correlation total | 18188.448 | 15 | ||||
| R-squared | 0.9692 | |||||
| Adj. R-squared | 0.9423 | |||||
| Pred. R-squared | 0.8769 | |||||
| Adeq precision | 17.3087 | |||||
*DF: Degree of freedom.
Estimated coefficient values for the parameter and parameter interaction effects
| Coefficient estimate | DF | Standard error | |
|---|---|---|---|
| Intercept | 43.148 | 1 | 2.091 |
| A | 3.444 | 1 | 2.091 |
| B | −10.820 | 1 | 2.091 |
| C | −28.173 | 1 | 2.091 |
| D | 8.745 | 1 | 2.091 |
| AC | −6.335 | 1 | 2.091 |
| AD | −5.606 | 1 | 2.091 |
| BC | 5.575 | 1 | 2.091 |
The analysis was conducted considering the predictability of the model at 95% confidence, implying that factors with a p-value < 0.05 have significant effects on the response. Based on the p-values as obtained by ANOVA (Table 5), the factors B (initial chromium concentration), C(pH), D (time) and the interaction terms AC (T°–pH), AD (T°–time) and BC (initial chromium concentration–pH) were found to be statistically highly significant. The high p-value ( >0.05) of A (T°) (0.1381) indicated the insignificance of this term in chromium degradation.
The analysis also revealed the high significance of the model which is evident from the F-value (36.00) with a low probability value (p < 0.0001) indicating that the first-order polynomial equation was statistically significant and can be used for prediction of chromium removal. The R2 (coefficient of determination) value of 0.969 and the adjusted R2 of 0.942 were close to 1.0 proving that the model shows a high correlation between the experimental and the predicted values. In addition, R2 and adjusted R2 are close to each other indicating that the model does not include insignificant variables [50]. This can be explained by the fact that the addition of significant independent variables to the model will increase adjusted R2 value while addition of nonsignificant ones will result in decreasing adjusted R2, whereas the R2 value will increase regardless of the added variable’s significance. Alternatively, the R2 value indicates that the proposed first-order polynomial regression model could explain 96.9% of the total variations in the biodegradation of Cr(VI). In addition, the predicted R2 was 87.7% suggesting that the predicted response could be well calculated by the model. The standard error of the mean (SEM) allowed us to determine the standard deviation of the residuals with a value equal to 2.091.
Thereby, ANOVA results clearly showed a linear relationship between all the main effects and the response. The significant predictive equation for chromium removal percentage is given below:
| (2) |
The obtained results were confirmed by the Pareto chart (Figure 2) also used for checking the significance of factors by displaying the t-values of the effects which are proportional to their degree of significance. The values of the effects were calculated by the formula and compared with reference lines: standard t-value = 2.26216 and the more rigorous Bonferroni limit = 3.95422. According to Myers et al. [52], all the effects above the Bonferroni limit are certainly significant to very important, while the ones above the standard t-value limit are possibly significant to moderately important. The Pareto chart for chromium removal by S10C1 strain (Figure 2) revealed that B and C are factors with the highest significance. Therefore, D, AC, AD and BC factors may be considered as moderately important. The sequence of the significant main effects is found to be C > B > D > AC > AD > BC.
Pareto chart of effects.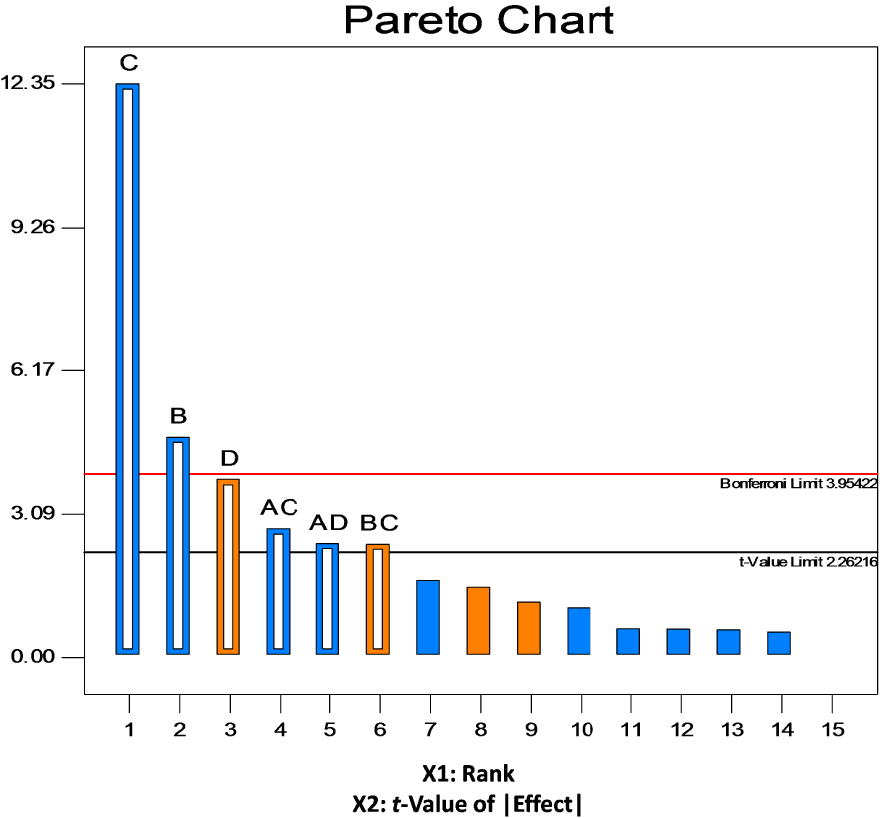
Correlations between experimental and predicted responses.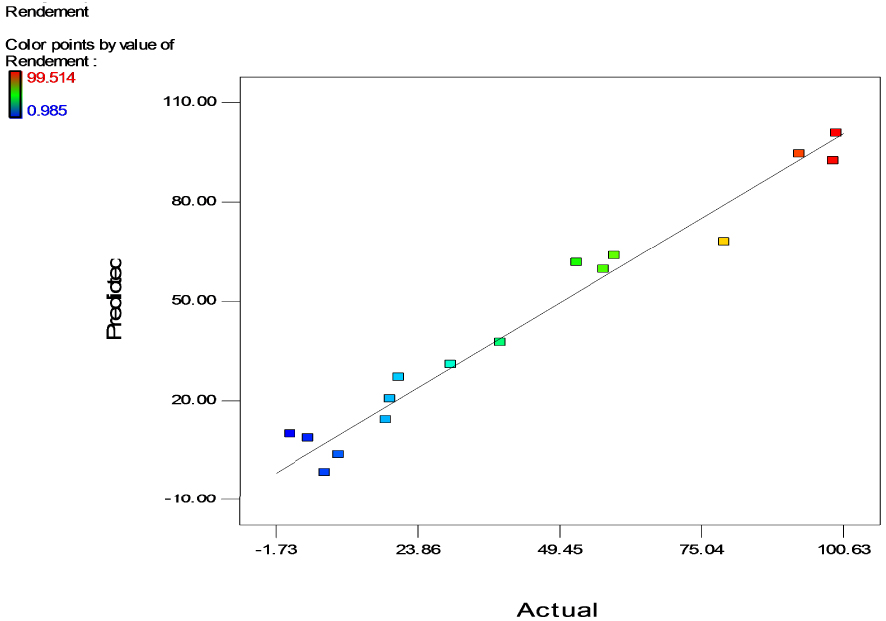
Figure 3 represents the relationship between the experimental values of Y and the predicted values obtained from the mathematical model. The residuals for majority of the responses are close to the straight line in the plot indicating a good match between the observed and the predicted values which means that both values are accurate and reliable and confirms the good prediction of the model. The good correlation between response and the fitted model, observed in the current study, was also demonstrated by other researchers [53, 54, 55, 56] who used a quadratic model to predict the maximum Cr(VI) removal by bacteria and confirmed that it appropriately explained the effects of the investigated parameters on chromium removal.
3.4.2. Interaction effects of variables
The relationships between the four variables (T°, initial chromium concentration, pH and time) and their interaction effects were analyzed using the 3D response surface plots. The 3D plots were created by plotting Cr removal% (response) on Z-axis and two independent variables on X-axis and Y -axis while keeping the other variables at fixed level (Figure 4).
The 3D response surface plots and their corresponding contour plots showing the effect of (a) pH and temperature (b) temperature and time (c) pH and initial chromium concentration on chromium degradation.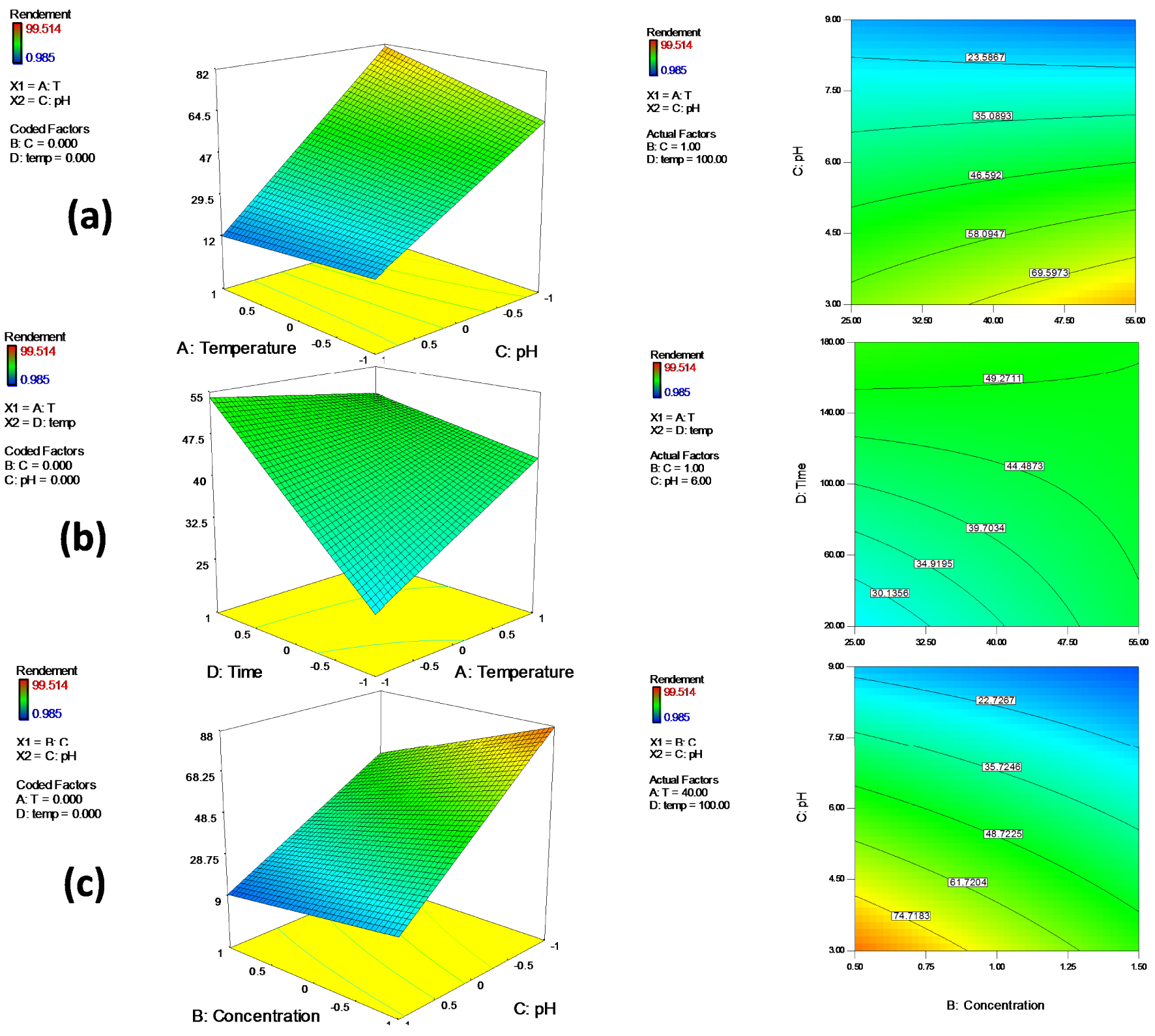
The interaction effect between pH and temperature is displayed in Figure 3a. As can be clearly observed, the chromium removal yield increases with decreasing pH to 3 and increasing temperature that was in agreement with optimal values obtained by solving the polynomial equation. This result can be explained by the fact that high pH values lead to the formation of metal complexes which decrease the concentration of chromium ions and then cause a decrease in the removal rate. The interaction effect of time and temperature is revealed in Figure 3b, the maximum Cr(VI) removal rate was recorded with increasing time and temperature. Figure 3c shows the combined effect of pH and initial chromium concentration. In this figure, maximum biodegradation capacity was obtained with decreasing pH and initial Cr(VI) concentration. These plots clearly show that pH–temperature interaction was the most significant factor that affected the chromium removal yield by the bacterial strain S10C1. Also pH interacted strongly with Cr(VI) concentration indicating predominant influence on Cr(VI) removal. Similar findings were reported by other researchers [57, 58, 59, 60, 61] who found that since the absorption of chromium by bacteria is mediated by proteins and enzymes, any change in pH and temperature may affect the enzyme ionization degree and the protein folding, consequently affecting enzyme activity and chromium reduction. It can be concluded that an acidic initial medium is beneficial for the bacterial strain S10C1 to achieve the highest chromium removal rate. Optimal pH and temperature for Cr(VI) elimination may be different from one microorganism to the other [62].
3.4.3. Optimization of variables
The objective of this step was to find the optimal experimental conditions, including temperature, pH, Cr(VI) initial concentration and retention time that lead to a maximum yield of chromium removal. This was achieved using a multiple response method called desirability function. The Design-Expert software, equipped with a response optimizer feature, generates a list of optimized yields with all possible combinations of the analyzed factors set at desired levels [63].
Comparison of chromium removal percentage with some microorganisms
| Microorganisms | % removal | Reference |
|---|---|---|
| Acinetobacter species | 99.95 | [56] |
| Aspergillus species | 92 | [31] |
| Micrococcus species | 90 | |
| A. fumigatus, A. flavus and A. fumigatus (Microbial consortium) | 81.25 | [44] |
| B. brevis | 77.24 | [64] |
| B. cereus | 59.28 | [54] |
| B. subtilis and B. safensis (co-cultured microcosm) | 95 | [46] |
| B. subtilis | 88 | |
| B. safensis | 91 | |
| Bacillus subtilis strain SZMC 6179J | 93.50 | [53] |
| Bosea sp. | 98 | [65] |
| Pseudomonas alcaliphila NEWG-2 | 96.60 | [55] |
| B. cereus strain S10C1 | 92.86 | Present study |
Maximum chromium removal of 94.4% was obtained with S10C1 strain at a temperature of 55 °C, an initial Cr(VI) concentration of 0.5 mM, a pH of 3 and a contact time of 20 h with a desirability value of 0.944 which is very close to the ideal desirability d = 1.0000. Under these conditions, the experimental removal yield was found to be equal to 92.9% in good agreement with the calculated one. This closeness between the experimental and the predicted values demonstrates the success of the Full Factorial Design, the correct selection of factors involved in the design as well as the accuracy of the adopted linear regression model.
The above results are comparable with those obtained by Liu et al. [53] on Bacillus subtilis strain SZMC 6179J using Box–Behnken experimental design, the predicted optimum Cr(VI) removal rate was 93.5% and this was confirmed experimentally. The study also revealed that pH was the most significant factor on chromium removal which was in accordance with our results. The Box–Behnken design was also used for the optimization of Cr(VI) adsorption on bacterial strain Bacillus brevis [64], maximum removal of Cr(VI) (77.2%) was achieved at pH 2.0, a temperature of 40 °C and initial chromium concentration 35 mg⋅L−1. The most significant factor was pH with a negative effect on Cr(VI) biosorption followed by temperature with a positive effect and then initial concentration of Cr(VI) which showed negative effect. The antagonistic effect of pH and initial concentration of Cr(VI) and the synergistic effect of temperature were also observed in the present study. Other investigations on dried Bacillus cereus strain using central composite design reported a predicted value of the adsorbed amount of Cr(VI) of 30.93 mg⋅g−1 which is very close to the experimental value of 31.34 mg⋅g−1 [54]. These authors indicated the significant impact of biomass dosage among the studied factors (initial Cr(VI) concentration, pH and biomass dosage).
Compared to other microorganisms, the maximum Cr(VI) removal efficiency of Pseudomonas alcaliphila NEWG-2 using face-centered central composite design (FCCD) was found to be 96.33% which was experimentally verified (96.60%) [55]. The optimized parameters were yeast extract (5.6 g⋅L−1) and glucose (4.9 g⋅L−1) as nutritional variables, pH (7) and incubation period 48 h as physical variables. Likewise, bacterial strain identified as Bosea sp. showed maximum removal efficiency of Cr(VI) of 98% using central composite design of response surface methodology [65], the optimum conditions were determined to be pH of 2, initial Cr(VI) concentration of 55 mg⋅L−1 and biomass dose of 2.0 g⋅L−1. Mrudula et al. [56] used Plackett–Burman design and response surface methodology to optimize factors controlling Cr(VI) removal by Acinetobacter species. Maximum Cr(VI) bioreduction percent was about 99% closely related to the experimental result of 99.95%.
In view of these results, it can be inferred that these statistical approaches were found to be effective in screening the most significant parameters as well as their interaction effects on chromium removal and on improving the efficiency of the process by a variety of bacteria, consistent with results from the current study. Additionally, as can be seen from Table 7, Bacillus cereus 4080 LBK, selected in the present work, showed a clear advantage with regard to chromium reducing efficiency over other bacterial strains, particularly the bacillus species.
4. Conclusion
In the present study, a chromium resistant bacterial strain S10C1, identified as Bacillus cereus 4080 LBK, was isolated from contaminated soil and sludge and investigated for its efficiency in Cr(VI) removal. For this purpose, a statistical approach based on a 24 full factorial experimental design was used for optimization of the operational variables including pH, initial Cr(VI) concentration, temperature and contact time. The maximum Cr(VI) removal efficiency of Bacillus cereus 4080 LBK was found to be 94.4% which was 33% higher as compared to unoptimized conditions. Under the optimal conditions, the experimental Cr(VI) removal percentage was evaluated at 92.9% which is close to the predicted value by the statistical design (94.4%). Thus, the use of full factorial design was found to be a suitable statistical tool in improving Cr(VI) removal efficiency and standardizing optimum conditions.
The indigenous bacterium Bacillus cereus investigated has demonstrated high capacity of resistance and reduction of chromium in extreme conditions such as at very low pH, high temperature and a relatively short time which makes it an interesting candidate for efficient Cr(VI) bioremediation in contaminated sites.
Further investigations are needed to confirm the potential of the Bacillus cereus 4080 for the bioremediation of chromium pollution, we cite specially:
- Further characterization of the strain to elucidate the mechanism of the process.
- Study of all factors affecting the process as soil characteristics and presence of other pollutants.




 CC-BY 4.0
CC-BY 4.0