Version française abrégée
La girelle, Coris julis (Linnaeus 1758 ; Pisces : Labridae) est un poisson marin côtier, sédentaire à l'âge adulte, mais qui présente une phase larvaire planctonique de quatre à six semaines. Elle possède également une grande aire de répartition. Cette espèce constitue donc un bon modèle d'étude de la différenciation génétique en milieu marin et de l'impact de la phase larvaire sur les flux de gènes. Une analyse génétique devrait aussi préciser les capacités de dispersion de cette espèce. D'autre part, les populations de girelles présentent des différences morphologiques entre le Cap-Vert et les autres populations, ainsi qu'entre Atlantique et Méditerranée. Nous avons cherché à déterminer la signification biologique de ces variations : s'agit-il de différences génétiques ou d'un cas de plasticité phénotypique ? La variabilité génétique de plusieurs populations de Coris julis a été analysée à différentes échelles géographiques pour tenter de répondre aux questions suivantes.
- (i) Les différences morphologiques observées correspondent-elles à des entités génétiquement différenciées ? Pour cela nous avons étudié et comparé des populations atlantiques (y compris du Cap-Vert) et méditerranéennes morphologiquement distinctes.
- (ii) Quel est le niveau de différenciation génétique et donc de flux de gènes entre populations insulaires (Açores) et continentales (péninsule Ibérique) ? La colonisation de cet archipel s'est-elle accompagnée d'un effet fondateur ?
- (iii) À une échelle locale, quelles sont les relations génétiques entre populations proches : entre ı̂les des Açores et le long de la côte atlantique de la péninsule Ibérique ?
Huit locus microsatellites, développés pour cette espèce, ont été appliqués à trois échantillons des Açores, deux du Portugal continental, un du Pays Basque, un de l'archipel du Cap-Vert, un de Madère, et quatre de Méditerranée. Certains échantillons ne possédaient qu'un faible nombre d'individus (Madère, Cap-Vert, échantillons méditerranéens) et l'interprétation des résultats a tenu compte de l'existence possible de biais d'échantillonnage. Les données obtenues ont été utilisées pour tester l'adéquation des populations à l'équilibre de Hardy–Weinberg et analyser les différences génétiques entre populations.
Le locus F2-4 présentait des déficits en hétérozygotes généralisés liés à la difficulté de détermination de la taille des fragments amplifiés et il n'a donc pas été utilisé pour les tests d'écart à l'équilibre de Hardy–Weinberg. Les sept autres locus ne présentaient aucun déséquilibre marqué et le niveau de variabilité obtenu était bien adapté à nos problématiques. La plupart des échantillons étaient à l'équilibre de Hardy–Weinberg. Seule la population du Pays basque s'en écartait ().
En ce qui concerne l'échantillon du Cap-Vert, seuls quatre individus ont été analysés. Néanmoins, la présence d'allèles privés pour cette population et l'absence de produits d'amplification par PCR pour un des locus indiquent bien l'originalité génétique de ces individus par rapport aux autres populations. Ce résultat est en accord avec les différences morphologiques et avec le fort taux de divergence de séquence de l'ADN mitochondrial observé dans une étude précédente. Ces données vont donc dans le sens d'un statut spécifique de ces populations (Coris atlantica). Ce statut avait été initialement proposé par Günther en 1862.
La comparaison des populations atlantiques (hors Cap-Vert) et méditerranéennes de Coris julis a mis en évidence l'existence de différences génétiques marquées de part et d'autre du détroit de Gibraltar. Même si ces résultats doivent être pondérés par la plus faible taille des échantillons méditerranéens, ils sont confortés par la présence d'allèles privés en Méditerranée, le positionnement des individus dans une analyse factorielle des correspondances (AFC), ainsi que par les Fst significativement différents de 0 entre populations des deux groupes. Les divergences de séquence de l'ADN mitochondrial vont dans le même sens. Toutes ces données indiquent une restriction aux flux de gènes au niveau du détroit de Gibraltar. L'existence d'une différenciation génétique entre Atlantique et Méditerranée a déjà été observée pour d'autres espèces marines et confirme le rôle de barrière biogéographique joué par ce détroit. Par ailleurs, dans le cas de Coris julis, l'existence de différences morphologiques correspond donc à la présence de deux entités génétiquement différenciées. Une analyse plus complète sera nécessaire pour préciser la structure génétique en Méditerranée.
L'analyse des échantillons atlantiques n'a pas permis de mette en évidence de différences génétiques significatives entre les populations des Açores et celles de la péninsule Ibérique, à l'exception du Pays basque. Le regroupement des individus des Açores et du Portugal continental constitue un échantillon en équilibre de Hardy–Weinberg (). Ces données indiquent donc la possibilité de flux de gènes réguliers entre le continent et l'archipel malgré plus de 1800 km de distance a priori sans habitat disponible et avec des courants défavorables. Ces échanges sont probablement liés à la durée de la phase larvaire et favorisés par des conditions océanographiques exceptionnelles. Par ailleurs, nos résultats n'ont pas montré l'existence d'un effet fondateur lié à la colonisation des Açores. Au sein de l'archipel des Açores, aucune différence génétique n'a été mise en évidence entre les différentes ı̂les. En ce qui concerne la péninsule ibérique, l'échantillon du Pays basque se différencie des deux populations portugaises analysées. Le mode de différenciation génétique le long de la côte ibérique devra être précisé par l'analyse d'un plus grand nombre d'échantillons.
La synthèse des données microsatellites et de celles issues du séquençage de l'ADN mitochondrial (ARNr 12S ; résultats d'une étude précédente) permet de proposer un scénario évolutif au cours duquel Coris atlantica et C. julis auraient divergé il y a 4 à 8 Ma. La divergence Méditerranée/Atlantique serait plus récente (1–2 Ma).
1 Introduction
As they present many different reproductive strategies and life histories, marine fish are good models to study genetic differentiation in an open environment (the sea). For species with high dispersal ability and/or long larval stages, one should theoretically observe low levels of genetic differentiation. Nevertheless, genetic studies have shown that more or less pronounced genetic differentiation can take place between seas or oceans, as for example concerning the relationships between the Atlantic Ocean and the Mediterranean Sea populations [1]. This can even be observed for species with high dispersal abilities such as for instance tuna [2] or other species (review in [3]). Significant genetic differences can also appear within a given sea [3–5].
An increasing number of studies on marine fish genetic structure are available, especially concerning exploited species. Nevertheless, the degrees of isolation of marine communities of rather isolated north-Atlantic islands, such as the Azores, remain to be studied [6], though it constitutes an interesting model of colonisation processes. The coastal species of these volcanic islands must have colonised this area from continent or other islands. Moreover, the different islands of the archipelago provide a good opportunity to study population structure at different spatial scales (among more or less proximate islands, between archipelago and continental populations).
The rainbow wrasse, Coris julis (Linnaeus 1758; Pisces: Labridae) is a small rocky shore fish that inhabits shallow waters and is site-attached throughout its adult life. It should then present few dispersal abilities in the adult stage. Its repartition area is fragmented among islands or between archipelago and continent. For example, there is apparently no suitable habitat for this species between European continent and Azores. It does however have pelagic eggs and a planctonic larval stage of four to six weeks. Thus, this species can be used as a model to study the consequences of larval stage on gene flow in a more or less fragmented habitat. Moreover, this species lives in a very large and heterogeneous geographical area (Mediterranean Sea, southern Black Sea, northeastern Atlantic from Sweden to south of Cape Lopez, Gabon, including the Azores, Madeira and the Canary Islands [7]) and presents conspicuous morphological variations between populations. Coris julis is a protogynous hermaphroditic species with diandry, i.e. with two types of males: primary males and secondary males, the later arising from females by sex change (see [8] for a review on this species biology). Secondary males present a colour pattern very different from the one common to females and primary males. Moreover, secondary males from the Mediterranean Sea and the Atlantic Ocean are differently coloured [9,10]. Several colour patterns can be observed in the Mediterranean Sea in the same location but at different depths [8,10]. Colour and morphological variations were also observed for fish from Sierra Leone (Gulf of Guinea), which were considered by several authors as another example of polymorphism inside the Coris julis species [11–13], although they had been originally described as a distinct species (Coris atlantica) by Günther [14].
In the present report, we studied the genetic relationships between different C. julis populations, in order to describe the genetic structure of this species at different scales and to answer the following questions.
- (i) At a morph scale: what are the genetic relationships among Atlantic and Mediterranean populations? In other words, do the morphological differences correspond to genetically differentiated units or to phenotypic plasticity? Moreover, can we confirm the specific status of Cape Verde individuals?
- (ii) What are the relationships among Atlantic C. julis populations, and more specifically, what are the genetic relationships between continental and insular populations of this species? We would expect high genetic divergence because of the lack of suitable habitats for this species between the continent and the Azores (distance: 1800 km without islands). The genetic relationships between insular and continental populations would be helpful to infer the colonisation history and process of this species in the Azores. Moreover, we can test whether this colonisation was linked with founder effects.
- (iii) At a local scale, what is the pattern of genetic structure between neighbouring populations? Can we detect local differentiation within the Azores archipelago or along the European coasts? For this coastal species, we would expect an isolation by distance model along continental coasts and maybe another differentiation model among islands depending on the dispersal abilities.
For this purpose, we chose to analyse samples originating from the Azores and several places along the continental coasts of Portugal and Spain. Other populations from the Cape Verde islands, Madeira and the Mediterranean Sea were also analysed. Samples sizes of these last locations were small, we will then present preliminary results and statistical parameters should be considered cautiously.
Such a study requires a priori neutral genetic markers variable enough for local scale studies and which can also be applied to ethanol-preserved fish or pieces of fins (more convenient for samples collection), such as microsatellites [15]. Microsatellites loci are very useful for discriminating populations with low genetic differentiation (see for example [16]) and can sometimes evidence population structure where other markers indicated an apparent homogeneity [17]. Moreover, microsatellite loci have already been specifically developed for this species [18]. We present here the first results concerning the genetic structure of C. julis, as determined from the analysis of eight microsatellite loci.
2 Materials and methods
2.1 Sampling and DNA extraction
Two hundred and forty-nine individuals were analysed. The sampling scheme, dates, locations and sample sizes are given in Table 1 and Fig. 1. We analysed three populations from the Azores (Faial, Corvo and Santa Maria) and three from the Iberian coasts (Faro, Arrábida and Basque Country), representing 204 individuals. We also analysed other samples originating from Atlantic (Cape Verde and Madeira) and Mediterranean Sea with smaller samples sizes (2 to 18 individuals). In the Azores, the Faial Island was sampled at two different sites and dates in order to test for an intra-island differentiation and temporal stability; as these two samples did not appear differentiated (see Results section), they were grouped in Table 1. Most samples consisted on pieces of fins preserved in ethanol. DNA preparation was performed using a standard phenol-chloroform extraction protocol or using Chelex 100 resin (Bio-Rad, Hercules, CA) [19].
Allelic frequencies of the different populations of C. julis used in this study
| Continent | Azores | Mediterranean Sea | ||||||||||
| Faro | Arrábida | Basque | Faial | Corvo | Santa | Cape | Madeira | Crete | Ibiza | Ligure | Marseille | |
| country | Maria | Verde | ||||||||||
| N | 27 | 30 | 30 | 62 | 22 | 33 | 4 | 3 | 2 | 5 | 13 | 18 |
| D-11 | ||||||||||||
| 88 | 0 | 0 | 0 | 0.02 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0 |
| 92 | 0 | 0 | 0 | 0.02 | 0.07 | 0 | – | 0.33 | 0 | 0.10 | 0 | 0 |
| 96 | 0.13 | 0.10 | 0.12 | 0.11 | 0.19 | 0.09 | – | 0 | 0 | 0.10 | 0.12 | 0.31 |
| 100 | 0.30 | 0.42 | 0.33 | 0.29 | 0.26 | 0.32 | – | 0 | 0 | 0.20 | 0.38 | 0.22 |
| 101 | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0.04 | 0.03 |
| 104 | 0.31 | 0.25 | 0.22 | 0.28 | 0.19 | 0.32 | – | 0 | 0 | 0.40 | 0.12 | 0.14 |
| 108 | 0.15 | 0.15 | 0.20 | 0.19 | 0.24 | 0.18 | – | 0.33 | 1 | 0.20 | 0.19 | 0.14 |
| 112 | 0.07 | 0.05 | 0.08 | 0.06 | 0.05 | 0.08 | – | 0.17 | 0 | 0 | 0.12 | 0.11 |
| 116 | 0.02 | 0.02 | 0.05 | 0.03 | 0 | 0 | – | 0.17 | 0 | 0 | 0.04 | 0.03 |
| 120 | 0.02 | 0 | 0 | 0.01 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0.03 |
| 124 | 0 | 0.02 | 0 | 0 | 0 | 0.02 | – | 0 | 0 | 0 | 0 | 0 |
| D3-3 | ||||||||||||
| 100 | 0.56 | 0.67 | 0.76 | 0.66 | 0.64 | 0.53 | 0 | – | 0 | 0.90 | 0.88 | 0.92 |
| 102 | 0.41 | 0.30 | 0.24 | 0.31 | 0.34 | 0.47 | 0 | – | 1 | 0.10 | 0.12 | 0.08 |
| 104 | 0.04 | 0.03 | 0 | 0.02 | 0.02 | 0 | 1 | – | 0 | 0 | 0 | 0 |
| 6-1 | ||||||||||||
| 98 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 99 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 100 | 0.91 | 0.93 | 0.93 | 0.90 | 0.84 | 0.89 | 0 | 1 | 1 | 0.80 | 0.77 | 0.92 |
| 101 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 102 | 0.09 | 0.07 | 0.05 | 0.07 | 0.16 | 0.11 | 0 | 0 | 0 | 0.20 | 0.08 | 0.06 |
| 104 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.15 | 0.03 |
| A-1 | ||||||||||||
| 91 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0.25 | 0 | 0 | 0 | 0 | 0 |
| 98 | 0.15 | 0.13 | 0.25 | 0.24 | 0.16 | 0.21 | 0 | 1 | 0 | 0 | 0 | 0 |
| 100 | 0.33 | 0.38 | 0.40 | 0.47 | 0.48 | 0.47 | 0.25 | 0 | 0 | 0 | 0 | 0.08 |
| 101 | 0.11 | 0.10 | 0 | 0.06 | 0.14 | 0.08 | 0 | 0 | 0 | 0 | 0 | 0 |
| 102 | 0.41 | 0.38 | 0.31 | 0.22 | 0.23 | 0.21 | 0 | 0 | 0 | 0.10 | 0.15 | 0 |
| 103 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| 104 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0.25 | 0 | 0.50 | 0.30 | 0.46 | 0 |
| 105 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.39 |
| 106 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.50 | 0.30 | 0.31 | 0.06 |
| 107 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.36 |
| 108 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0.11 |
| 110 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 112 | 0 | 0 | 0.02 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0.30 | 0.08 | 0 |
| F-7 | ||||||||||||
| 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.25 | 0.30 | 0.31 | 0.42 |
| 96 | 0 | 0.07 | 0 | 0.02 | 0.02 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 |
| 98 | 0.33 | 0.28 | 0.10 | 0.38 | 0.43 | 0.29 | 0 | 0.33 | 0.50 | 0.40 | 0.50 | 0.33 |
| 99 | 0 | 0 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 100 | 0.44 | 0.45 | 0.50 | 0.42 | 0.34 | 0.52 | 0 | 0 | 0 | 0.10 | 0 | 0 |
| 102 | 0.10 | 0.08 | 0.25 | 0.03 | 0.09 | 0.05 | 0 | 0.33 | 0.25 | 0.20 | 0.12 | 0.11 |
| 104 | 0.10 | 0.10 | 0.04 | 0.08 | 0.07 | 0.09 | 0 | 0.33 | 0 | 0 | 0.08 | 0.14 |
| 106 | 0.02 | 0.02 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 108 | 0 | 0 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 110 | 0.02 | 0 | 0 | 0.04 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| 112 | 0 | 0 | 0 | 0.01 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 116 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 120 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 122 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 126 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 128 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 134 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 136 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 144 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 150 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 162 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| G2-2 | ||||||||||||
| 82 | 0 | 0.02 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 94 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0.03 |
| 96 | 0 | 0.02 | 0 | 0.01 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0.03 |
| 100 | 0.09 | 0.07 | 0.15 | 0.14 | 0.07 | 0.14 | 0 | 0 | 0 | 0.10 | 0.04 | 0.08 |
| 102 | 0.85 | 0.82 | 0.81 | 0.82 | 0.88 | 0.83 | 0 | 0.75 | 0.50 | 0.60 | 0.69 | 0.72 |
| 104 | 0 | 0.03 | 0 | 0.02 | 0 | 0.02 | 0.13 | 0 | 0.25 | 0.20 | 0.19 | 0.11 |
| 106 | 0.02 | 0.02 | 0.02 | 0.01 | 0 | 0.02 | 0.25 | 0 | 0 | 0 | 0.04 | 0 |
| 108 | 0.02 | 0.02 | 0 | 0 | 0.02 | 0 | 0.13 | 0.25 | 0.25 | 0.10 | 0 | 0.03 |
| 110 | 0 | 0 | 0 | 0 | 0 | 0 | 0.25 | 0 | 0 | 0 | 0 | 0 |
| 116 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 120 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 124 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 128 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F2-4 | ||||||||||||
| 1 | 0.15 | 0.17 | 0.04 | 0.07 | 0 | 0.20 | 0 | 0.25 | 1 | 1 | 1 | 1 |
| 2 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0.37 | 0.23 | 0.28 | 0.25 | 0.32 | 0.30 | 0 | 0.25 | 0 | 0 | 0 | 0 |
| 4 | 0.46 | 0.53 | 0.68 | 0.67 | 0.68 | 0.50 | 1 | 0.50 | 0 | 0 | 0 | 0 |
| 5 | 0.02 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E-4 | ||||||||||||
| 80 | 0 | 0 | 0 | 0 | 0 | 0 | 0.50 | 0 | 0 | 0 | 0 | 0 |
| 83 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 88 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 90 | 0.42 | 0.42 | 0.48 | 0.38 | 0.45 | 0.42 | 0 | 0.50 | 0.50 | 0.20 | 0.54 | 0.53 |
| 91 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 92 | 0.02 | 0.03 | 0.02 | 0.01 | 0.02 | 0.02 | 0 | 0 | 0 | 0.10 | 0 | 0.06 |
| 94 | 0 | 0 | 0 | 0.01 | 0.05 | 0.02 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 96 | 0 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 100 | 0.08 | 0 | 0.08 | 0.06 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 |
| 102 | 0 | 0 | 0.04 | 0.02 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0.06 |
| 104 | 0.04 | 0.07 | 0.04 | 0.06 | 0 | 0.02 | 0 | 0 | 0 | 0.10 | 0 | 0.03 |
| 105 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 106 | 0.02 | 0.02 | 0.04 | 0.05 | 0.07 | 0.06 | 0.25 | 0 | 0 | 0 | 0 | 0.03 |
| 108 | 0 | 0.02 | 0.02 | 0.03 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| 109 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0 |
| 110 | 0.02 | 0.05 | 0 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0.10 | 0 | 0.06 |
| 112 | 0.02 | 0.02 | 0 | 0.03 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0.03 |
| 114 | 0 | 0.02 | 0.02 | 0.02 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 116 | 0.02 | 0 | 0 | 0.02 | 0.02 | 0.05 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 118 | 0 | 0 | 0 | 0.03 | 0 | 0.02 | 0 | 0 | 0.25 | 0 | 0 | 0 |
| 120 | 0 | 0 | 0.02 | 0.03 | 0.02 | 0.02 | 0 | 0 | 0 | 0.10 | 0 | 0 |
| 122 | 0 | 0.02 | 0.02 | 0.03 | 0.02 | 0.02 | 0 | 0.25 | 0 | 0 | 0 | 0.08 |
| 124 | 0.02 | 0.03 | 0.02 | 0.01 | 0.05 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0.08 |
| 126 | 0.02 | 0.05 | 0 | 0.03 | 0.02 | 0.05 | 0 | 0 | 0 | 0.10 | 0.08 | 0 |
| 128 | 0.08 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 130 | 0 | 0.02 | 0 | 0.03 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| 132 | 0.02 | 0.02 | 0.02 | 0 | 0.02 | 0.03 | 0 | 0.25 | 0 | 0.10 | 0 | 0.03 |
| 134 | 0 | 0.05 | 0.02 | 0.03 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0.08 | 0 |
| 136 | 0 | 0 | 0.02 | 0.02 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 138 | 0.08 | 0.03 | 0.04 | 0.03 | 0.02 | 0.02 | 0 | 0 | 0.25 | 0 | 0 | 0 |
| 140 | 0.02 | 0.02 | 0 | 0.02 | 0 | 0.02 | 0 | 0 | 0 | 0.10 | 0.04 | 0 |
| 141 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 142 | 0 | 0 | 0 | 0.02 | 0.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 144 | 0 | 0 | 0.02 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0.04 | 0 |
| 146 | 0.02 | 0.02 | 0 | 0.02 | 0.05 | 0.03 | 0 | 0 | 0 | 0.10 | 0 | 0 |
| 148 | 0.06 | 0 | 0.02 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0.08 | 0.03 |
| 150 | 0 | 0 | 0.02 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 152 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 154 | 0 | 0 | 0 | 0 | 0.02 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| 156 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 158 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 160 | 0 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A moy | 6.1 | 7.3 | 6.1 | 8.5 | 5.6 | 6.8 | 3.7 | 2.4 | 1.9 | 3.9 | 4.5 | 5 |
| Hnb | ||||||||||||
| moy | 0.58 | 0.57 | 0.53 | 0.55 | 0.55 | 0.57 | 0.51 | 0.55 | 0.4 | 0.57 | 0.5 | 0.47 |
| Hobs | ||||||||||||
| moy | 0.56 | 0.53 | 0.42 | 0.52 | 0.48 | 0.52 | 0.54 | 0.57 | 0.5 | 0.63 | 0.5 | 0.46 |
| −0.03 | 0.03 | 0.15 | 0.02 | 0.06 | −0.01 | −0.06 | −0.23 | −0.45 | −0.10 | 0.01 | 0.02 | |
| 0.87 | 0.81 | 0∗ | 0.06 | 0.29 | 0.69 | 0.83 | 1 | 1 | 1 | 0.99 | 0.98 |
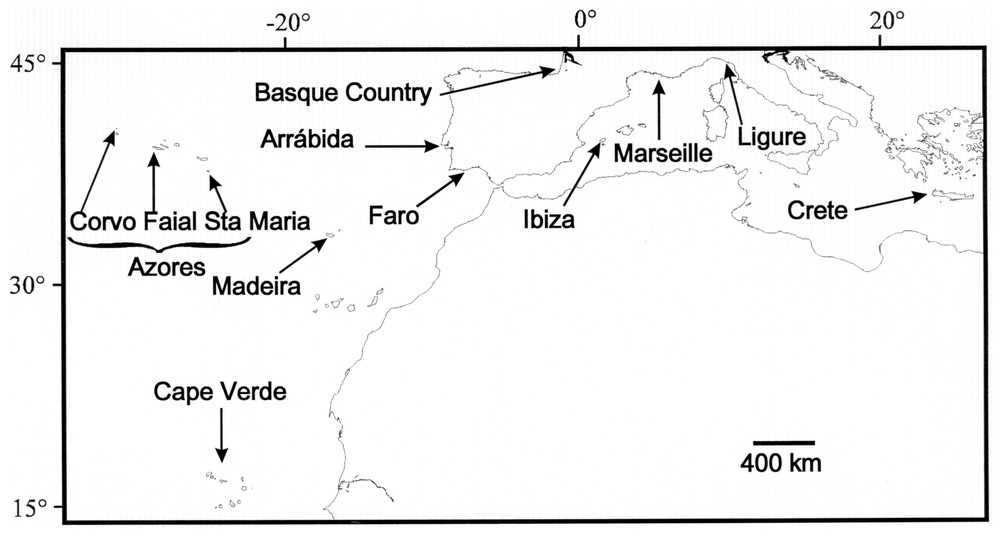
Location of the sampling sites. Longitudes (Greenwich, UK, as datum longitude) and latitudes are indicated at the top and the left of the frame.
2.2 Microsatellites analysis
Eight microsatellite loci have been used following the conditions already described [18]. Between one and three PCR products per individual were separated simultaneously (i.e. in the same lane) by electrophoresis on 6% polyacrylamide gels. For the F2-4 locus, the size of PCR products was too high to evaluate precisely the differences in number of repeats among the different alleles therefore this locus was not used to compute statistics taking into account allele sizes. The six observed alleles were numbered from 1 to 6, without any reference to their size.
2.3 Data analysis
Allelic frequencies and genetic diversity statistics (number of alleles, observed and expected heterozygosities) were computed using the GENETIX 4.0 software [20]. Genetic diversity within populations was evaluated by computing observed and unbiased expected heterozygosities [21]. Estimates of F statistic ( and ) [22] were also computed using GENETIX 4.0. Departure from Hardy–Weinberg equilibrium and genetic differentiation for each pair of populations were tested using the pseudo exact-test procedures implemented in the Genepop version 3.1 software [23]. Overall significance of several tests (for multi-locus analyses) was computed using Fisher's method [24]. Since the F2-4 locus presented a generalized departure from Hardy–Weinberg equilibrium across all populations (Fisher's method implemented in Genepop; p< 0.001), it was not used for the multi-locus tests of departure from Hardy–Weinberg equilibrium within each population.
In order to analyse the genetic differentiation between Atlantic and Mediterranean Sea Coris julis populations (Cape Verde excluded), we grouped Mediterranean samples on one side, and Atlantic ones on the other, and we realised a test of genic differentiation using GENEPOP.
When necessary, a sequential Bonferroni correction for multiple tests was applied [25].
In order to give an image of genetic relationships among individuals (and then partly avoiding some difficulties due to small samples sizes), a Factorial Correspondence Analysis (FCA) was done using Genetix. In this kind of analysis, individuals are first coded according to the presence of the different alleles with values 0 (does not possess the allele), 1 (heterozygote), 2 (homozygote for the allele). The computation then aims at finding composite axes (where each variable contributes differently to the global inertia), which optimise the differences between the analysed individuals. The inertia values along each axis can be assimilated into linear combinations of the monolocus Fst estimates [26]. As rare alleles can introduce much distortion in the FCA result, alleles present less than four times in the data set were not included in the analysis (as in [16]).
3 Results
3.1 Microsatellite variability
The numbers of alleles by locus and by population are given in Table 1. The total number of alleles by locus varied from 3 (locus D3-3) to 42 (locus E-4). No amplification products were obtained for Cape Verde individuals at the D-11 locus, which may be the consequence of primers sequence divergence or locus deletion.
Concerning departure from Hardy–Weinberg equilibrium, most loci showed no or moderate heterozygote deficits in most samples. Only the F2-4 locus presented high Fis values (p<0.001).
In the following parts, estimates of statistical parameters (such as F statistics) are presented for all samples. The analyses will focus on the largest samples: Azores and Atlantic continental samples (Faro, Arrábida and Basque Country). The small size of the other samples (Mediterranean, Madeira, Cape Verde) combined with the high variability of the microsatellites can lead to large variance of estimates. In these cases, population statistical parameters are given and discussed, but any interpretation will be made in the light of this possible bias.
3.2 Population variability
Allelic frequencies and multilocus estimations are given in Table 1. The mean expected heterozygosity varied between 0.53 (Basque Country) and 0.58 (Faro). The same level of heterozygosity was observed for continental and insular samples. Lower heterozygosities and number of alleles were observed for small samples (Mediterranean Sea, Madeira and Cape Verde).
When computed over these seven loci (F2-4 excluded), apart from Basque country, the populations showed none or little departure from panmixia. Accordingly, no significant linkage disequilibria were observed (data not shown). Particularly, when the two samples captured at Faial at different dates were grouped (as presented in Table 1), this did not lead to any significant departure from Hardy–Weinberg equilibrium. We then conserved this grouping for the other analyses. The Basque country population presented significant heterozygotes deficiency (), but with no generalised linkage disequilibria.
3.3 Azorean and continental Atlantic population differentiation
The between pairs of populations estimates [22] are given in Table 2. All pair wise comparisons are presented; nevertheless, those involving continental Atlantic and Azores samples are highlighted because these are the largest samples. For these populations, values varied between −0.01 and 0.01. The mean pairwise among these populations was 0.001 (SE=0.007). Among 15 comparisons, only four indicated a significant genetic differentiation at the 5% level after sequential Bonferroni correction (k=15). These significant concerned the comparisons between the Basque Country and the Azores, and between Basque Country and Faro. We did not find any evidence of genetic differentiation between other continental samples and Azores, nor among islands samples. An exact test of Hardy–Weinberg equilibrium performed over Atlantic C. julis populations, excluding Basque country and the small sample of Madeira, failed to reveal any departure from panmixia (global ). The same test performed over all populations, including Basque country, indicated a highly significant departure from Hardy–Weinberg equilibrium ().
Genetic differentiation between pairs of populations
| Cape Verde | Madeira | Faro | Arrábida | Basque | Faial | Santa Maria | Corvo | Ibiza | Ligure | Marseille | Crete | |
| country | ||||||||||||
| Cape Verde | – | 0.27 | 0.39 | 0.39 | 0.41 | 0.40 | 0.39 | 0.39 | 0.47 | 0.52 | 0.55 | 0.51 |
| Madeira | 0.078 | – | 0.08 | 0.09 | 0.06 | 0.08 | 0.08 | 0.07 | 0.18 | 0.21 | 0.20 | 0.25 |
| Faro | 0.002 | 0.012 | – | 0.16 | 0.19 | 0.20 | 0.24 | |||||
| Arrábida | 0.002 | 0.016 | – | 0.15 | 0.18 | 0.19 | 0.26 | |||||
| Basque country | 0.004 | 0.104 | – | 0.20 | 0.23 | 0.23 | 0.30 | |||||
| Faial | <0.001 | 0.02 | 0∗ | – | 0.19 | 0.21 | 0.22 | 0.27 | ||||
| Santa Maria | 0.003 | 0.031 | – | 0.17 | 0.20 | 0.21 | 0.23 | |||||
| Corvo | 0.006 | 0.041 | – | 0.19 | 0.22 | 0.23 | 0.26 | |||||
| Ibiza | 0.03 | 0.056 | <0.001 | 0.002 | 0.001 | <0.001 | 0.001 | <0.001 | – | 0.00 | 0.06 | 0.18 |
| Ligure | 0.014 | 0.012 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.575 | – | 0.07 | 0.18 |
| Marseille | 0.007 | 0.02 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.02 | <0.001 | – | 0.26 |
3.4 Comparisons with the other samples
All pairwise Fst values are given in Table 2, even those involving the smallest samples. When considering all samples, but excluding the Cape Verde sample (possibly corresponding to a different species), the mean was 0.133 (SE=0.099). When only considering the Atlantic populations, the mean pairwise was 0.025 (SE=0.036) with Madeira (three individuals), and 0.004 (SE=0.006) when Madeira was not included. For Mediterranean populations, the mean was 0.125 (SE=0.099) with Crete (two individuals) and 0.043 (SE=0.038) without Crete.
The corresponding pairwise tests mainly evidenced significant differentiation for the comparisons between Mediterranean and Atlantic populations.
The allelic frequencies observed for small Mediterranean samples were quite different from Atlantic ones and convergent patterns were observed for different populations. For example, Mediterranean samples exhibited some alleles absent or rare in Atlantic samples with high frequencies. At locus A-1, the 106 allele was present at frequencies higher than 0.3 in three samples and absent in Atlantic. The 104 allele presented frequencies varying from 0.3 to 0.5 in the same three Mediterranean samples, but was only present in one Atlantic individual. At the F2-4 locus, the 38 Mediterranean individuals analysed were homozygotes for the allele 1, which was not the most frequent one in Coris julis Atlantic (maximum frequency 0.17).
Concerning this locus, the probability of drawing 38 homozygous individuals for one allele present at a frequency of 0.17 (we choose the most favourable hypothetical situation, where the Mediterranean population would present the maximum frequency observed among the Atlantic ones) is (0.17)76. The mean frequency of this allele among Mediterranean samples necessary to get such an observation, with a probability higher than 0.05, is about 0.97. Similar computation can be made for other alleles, which indicates sharp differences among Atlantic and Mediterranean allelic frequencies.
In order to test for these differences, we grouped all Mediterranean populations together on one side (leading to a sample of 38 individuals), and all Atlantic Coris julis on the other side (207 individuals). These two groups were compared using a test of genic differentiation. The test was significant for each locus, and when combined over all loci (Fisher's method) gave an infinite Chi-2 value indicating a highly significant differentiation.
The Cape Verde sample exhibited high values when compared to other samples, but only one comparison indicated a significant genetic differentiation after a sequential Bonferroni correction. Several private alleles were found with only four individuals. For example concerning locus F-7, this populations exhibited eight private alleles (122 to 162) and no allele shared with other samples. Private alleles were also observed for locus 6-1, A-1, G2-2 and E-4.
3.5 Factorial correspondence analysis
The first FCA performed by taking into account the Cape Verde population (possibly corresponding to another species: C. atlantica) showed a large distance between these individuals and the other populations, but among C. julis samples, no clear relationship could be observed as they were grouped together (data not shown). A second FCA was then realised only with C. julis individuals. The result is shown in Fig. 2; it represents the projection of the individuals on the plane defined by the first two axes of the FCA. Axes 1 and 2 represented respectively 6.92% and 3.45% of total inertia. For this figure, all samples from Azores (three samples) were grouped together as they did not appear separated on this plan and were not genetically differentiated.

Results of the Factorial Correspondence Analysis. The individuals of the different samples (excluding Cape Verde, see text) are plotted on axes 1 and 2, which represented respectively 6.92 and 3.45% of total inertia. Individuals from Azores were grouped together.
The first axis of the FCA clearly separated Atlantic (on negative side, Fig. 2) and Mediterranean individuals. Concerning Atlantic individuals, the continental and Azores samples grouped together in a cloud with low extension, which indicated the genetic homogeneity of this group. Concerning Mediterranean Sea individuals, the samples appeared more differentiated and two main groups (Marseille and the other ones) seemed to be present and separated along axis 2. Within the two Mediterranean groups, the individuals appeared genetically more distinct from each other than within the Atlantic group, as suggested by the relative sizes of the different ‘clouds’. Axis 3 (3.24% of inertia) did not bring much more information and mainly separated three individuals from Faro, Arrábida and Basque country (data not shown).
4 Discussion
4.1 A relatively low microsatellite variability in C. julis populations
The eight microsatellite loci used here appeared well suitable for our study. Nevertheless, one of them (F2-4) showed high and generalized heterozygotes deficiencies ( estimates up to 0.77), which were not corroborated by the other loci. As already mentioned, this locus had alleles of large sizes, which were probably not well separated when they only differed from a few bp. Some of them most probably represent groups of alleles, and such a phenomenon can create heterozygote deficiencies. The observed apparent departures from panmixia could also be the consequence of the presence of some null alleles at this locus.
The Basque Country population was the only one presenting heterozygote deficiencies at several loci. The concomitant presence of null alleles for different microsatellites in this population only is not likely. A possible explanation would be a Wahlund effect, with the presence of several genetically differentiated entities in the same sample. The Factorial Correspondence Analysis did not evidence any sub-groups among these individuals and then we could not favour this hypothesis. Inbreeding is another possible cause of heterozygote deficiencies. If this were the case, it would mean that this population has a different social structure (and consequently different mating systems) than the other ones. No behavioural data are available at present to test this hypothesis.
The analysed populations presented quite high observed and expected heterozygosity values, typical of microsatellite loci. Nevertheless, these values are lower than usually observed for marine fish. A review concerning molecular markers cited expected heterozygosities (obtained with microsatellites) for marine fishes varying between 0.84 and 0.92 [27] (0.53 to 0.58 for C. julis). Coris julis would then be genetically less variable than other marine fishes or even many freshwater fishes [28]. This is probably linked with the biological characteristics of this species, which being sedentary and territorial may present lower effective population sizes than the pelagic species listed in the already cited paper [27]. This emphasizes the need to study other species with a variable range of site attachment. Nevertheless, the pelagic larval stage should enhance migration rate and then the genetic variability of the different populations. Protogynous hermaphrodite species may also present a lower genetic variability than dioecious ones because of a lower effective size [28]. Another explanation could be that this species experienced reduced population sizes in its history, which reduced its genetic variability.
4.2 The evolution of insular fish fauna: are the Atlantic populations a panmictic unit?
Among the Atlantic C. julis populations, only the Basque Country population appeared genetically different from the Azores and Faro samples, but not from Arrábida, which is the closest sample to the Basque Country. No genetic differentiation was observed between Azorean and other continental populations (Faro and Arrábida), despite more than 1800 km of distance between continent and this archipelago. These continental and insular populations could form a panmictic unit, as we did not evidence any departure from Hardy–Weinberg equilibrium. This is a surprising result when considering that the high degree of geographical isolation of the Azores islands makes recurrent gene flow between the continent and the islands a priori very unlikely [6]. This is especially true for coastal species, which do not have suitable habitats (i.e. shallow areas above 50 m in depth) between continental Portugal and the Azores that can serve as ‘step stones’ for migration. The exceptions are the seamounts ‘Ormond’ and ‘Gorringe’, southeast of continental Portugal but, even in this case, it would imply high dispersal abilities.
Our results indicate that the migration rate must be high enough to prevent genetic differentiation at large (continent-Azores) and small (inside archipelago) scales. These exchanges could be realised through the larval stage of this species [8]. It must be the main path for exchanges among Atlantic populations, probably taking advantage of exceptional favourable oceanic conditions that enable their transport under survival conditions [6]. One should note that the Azorean shallow water marine fauna is zoogeographically related to the eastern Atlantic even though the currents (Canary Current) mainly come from the west (see references in [29]), and this observation is consistent with our results on C. julis. For example, dispersal from Africa to Azores has been proposed in a brooding bivalve [30].
The lack of differentiation of C. julis in Atlantic Ocean could also be the consequence of a recent homogeneous colonization of this area. Nevertheless, even this explanation requires high gene flow between continental and insular populations to avoid any founder effect and to prevent any further differentiation due to genetic drift (which is more intense for small populations). One should also note that the Azores samples presented the same level of genetic variability than continental ones, indicating that founder effects probably did not occur along with the islands colonisation.
Among the Atlantic C. julis, only the Basque Country population appeared genetically separated (differing from the Azorean ones and from Faro but not from Arrábida). It is possible that the geographical position of the Basque Country (inside the Gascogne Gulf) population greatly reduces genetic exchanges toward Azores but not with proximate continental Portugal, as easily deducible from the global oceanic current patterns, that flow towards southeast from the Azores [6]. The analysis of more samples along the Iberian coasts would be interesting in order to test for a possible isolation by distance phenomenon (only the nearest population, Arrábida, did not appear differentiated from Basque Country).
The genetic status of Madeira remains to be analysed with more individuals.
4.3 C. atlantica and C. julis as two distinct species?
The different private alleles obtained with only four fishes from the Cape Verde islands clearly indicated the originality of these individuals. This finding is in agreement with a previous analysis on mtDNA variability [31], which showed a high sequence divergence between these two putative species. This is also concordant with morphological data [32] and would support the specific status of C. atlantica. West African Coris were originally described as a distinct species (C. atlantica) by Günther [14].
4.4 C. julis as an example of Mediterranean/Atlantic divergence
Despite their low sample size, the allelic characteristics of the Mediterranean samples indicate a differentiation between these populations and the Atlantic ones. It is noticeable to observe that some alleles, absent or rare in Atlantic populations, are quite frequent in several of the Mediterranean samples, even if the samples sizes could have influenced these results. Such convergent observations support the idea of an Atlantic/Mediterranean Sea differentiation. The genic differentiation test confirmed these observations. We also obtained significant differentiation tests between Atlantic and Mediterranean populations, apart from Crete, even if the obtained values may probably present a large variance.
The Factorial Correspondence Analysis also indicated the genetic distinctiveness of Mediterranean populations. The positioning of the individuals seem to indicate that genetic differentiation in Mediterranean Sea could be more pronounced than inside the Atlantic Ocean, which would agree well with the observed values. Nevertheless the small samples sizes did not provide a precise estimation of the level of genetic differentiation inside the Mediterranean Sea.
Our preliminary results concerning Mediterranean and Atlantic samples can be discussed in the light of previously published results (mtDNA and morphology). Concerning the analysis of mtDNA variation [31], a private haplotype was found for the two Mediterranean (Crete) individuals analysed, which reinforces our results. This distinction would be coherent with the differences in colour pattern observed between Mediterranean and Atlantic individuals [7,9,10]. This genetic distinction between Atlantic and Mediterranean populations has already been observed for other marine organisms [1]. Such consistent trend reveals the importance of the Gibraltar strait as an important biogeographic barrier, even for species with possible larval dispersal. As already suggested the lowering of sea level during glacial events could have reduced the living area of coastal species (including C. julis) and accentuated the barrier effect of bathymetric steps [1].
4.5 The biogeography of C. julis
From the results of the present paper and that based on 12s mitochondrial rDNA [31], we can propose a biogeographical scenario accounting for the observed data and try to date a divergence time using molecular information.
Estimations of divergence time based on DNA sequence divergence should be carefully considered, especially in the absence of a fossil record leading to the impossibility of calibrating the molecular clock. From the 12S mtDNA data [31], a substitution rate of about 0.5 to 1% per million years (i.e. a divergence rate of 1 to 2% per MY) as proposed by some authors [33,34] would lead to a divergence time between C. atlantica and C. julis of about four to eight million years. As the 12S evolves more slowly than protein coding genes [35] from which the estimation of the mitochondrial substitution rate has been inferred, our estimation should be considered as a lower bound of the probable divergence time. The same argument leads to an estimate of 1–2 million years for the divergence between C. julis from the Atlantic and the Mediterranean Sea.
The nuclear (microsatellites) and mitochondrial (12S rDNA) divergence could therefore correspond to the following scenario. The Mediterranean Sea dried out and was then recolonized from the Atlantic about 6 million years ago [36,37]. At this time, an eastern Atlantic Coris probably entered the Mediterranean Sea and then evolved into the species C. julis. In parallel, C. atlantica evolved in African coasts. About 1–2 million years ago (corresponding to the Mediterranean/Atlantic divergence of C. julis), C. julis went out from Mediterranean Sea via the Strait of Gibraltar, and colonized the temperate northeastern Atlantic. Adaptive evolution to the different environmental conditions under the latitudinal gradient could then have caused (before or after the Atlantic re-colonization of C. julis) the geographic segregation that is observed today between the two species in the Atlantic. A similar process was speculated to explain the distribution of the blennioid fish Tripterygion tripteronotus [38]. In this situation, the Atlantic populations would be a homogeneous and recent subset of the Mediterranean ones.
The present migration rate through the Gibraltar strait would be low enough to maintain the differentiation with Atlantic or selection would act differentially in these two environments to prevent migrants from reproducing within a new population. Some individuals corresponding to an ‘Atlantic’ phenotype have already been observed in sympatry in the Mediterranean Sea [8,10]. They could correspond to rare recent migration from Atlantic into Mediterranean Sea. This could be tested by using microsatellites and assignation methods (see for example [39]), on the corresponding samples (i.e. individuals with Atlantic coloration sampled in Mediterranean Sea).
5 Conclusions
The analysis of the genetic structure of the rainbow wrasse with microsatellites led to three main conclusions corresponding to our three main objectives (see introduction).
- (i) The observed morphological differences correspond to genetically differentiated units (Atlantic/Mediterranean) or even different species (Cape Verde).
- (ii) No genetic differentiation was observed among Atlantic populations from continent and Azores, with the exception of the Basque Country. This indicates that gene flow between the Azores islands and continental Europe is higher than previously thought, favouring the idea that European affinities of insular fauna are sustained by regular population exchanges.
- (iii) Among Atlantic neighbouring populations, and in particular in the Azores archipelago, no genetic structure was observed, indicating regular gene flow between localities. The possibility of an isolation by distance should nevertheless be tested along Iberian coasts using more samples.
The next step of this study will be to analyse other Atlantic populations in order to understand what are the limits of the panmictic population evidenced here. Concerning the Mediterranean Sea, a more precise study of the biogeography of C. julis in this area should provide additional information that is required in order to interpret the observed genetic structure.
Acknowledgements
We would like to thank Iñaki Artetxe Irueta, Giampietro Sara, Mireille Harmelin-Vivien and Jean-Georges Harmelin, Pedro Neves, Mafalda Freitas, Ricardo Araújo, and Robert Patzner for providing tissue samples. This work was supported by grants from the Portuguese Foundation for Science and Technology (FCT): Praxis BPD 4470/96, Praxis BPD 22076/99, Praxis BIC 1938/96, Praxis BIC 1930/96 and Praxis EMG 1957/95. The experiments complied with the current laws in Portugal.


