1 Introduction
The terminology used for plant pathology is wide and diverse. It is also sometimes ambiguous or even contradictory when compared with the terminology of animal pathology [1] or because of the use of the same words in different languages. A classic example of the latter is the term ‘virulence’, which refers, in a French plant pathology glossary, to the capability of a pathogen to invade a plant. In animal pathology and in ‘English’ plant pathology, the same word means the extent by which this capability can be quantified. Moreover, the large diversity of plant–pathogen interactions sometimes makes the understanding of the associated concepts even more difficult. As described below, the main concepts are linked to different levels of specificity and resistance types, and to well-defined quantitative and qualitative aspects. Although they help in understanding concepts, these distinctions are not likely to be irreversible. Following an overview of the diversity of plant pathology concepts, the aim of this review is to show that, based on numerous examples of recent experimental observations, classic distinctions can be questioned, at least at the cellular and molecular levels.
2 The diversity of plant pathology concepts
Although plants lack an immune system similar to mammals, they do resist most of the pathogens that live in the rhizosphere or in the phyllosphere. Disease is an exception, not the rule: plants are so-called ‘functionally nonhosts’ to most of their putative parasites and both partners share fundamental incompatibility. However, some pathogens are known to exhibit a total lack of specialization and infect a wide host range, as exemplified by most necrotrophic fungi or bacteria that kill colonized plant tissues. On the other hand, because of the occurrence of a mutual perception between plants and their pathogens, other parasites show some specificity and are restricted to a limited range of hosts, even to a single host. Narrow specificities are most often exhibited by biotrophic parasites that maintain the host's structural and physiological integrity. Interaction specificity, if it occurs, determines whether the plant resists the pathogen or the disease becomes well established [2].
2.1 Host specificity
This first specificity level occurs when a parasite species is able to invade only a single plant species. It explains why a plant that is susceptible to a given parasite is also resistant to most others and it allows the following types to be defined:
- – forma specialis (f.sp.) or varieties (var.) of fungi;
- – pathovars (pv.) or varieties (var.) of bacteria;
- – pathotypes or strains of viruses;
- – races, biotypes, pathotypes or strains of nematodes.
The resistance shown by a plant involved in a nonhost relationship is called ‘nonhost resistance’. It occurs in all plants against the majority of potentially invasive agents and is the most common type of resistance.
Although they have not been extensively studied, preformed defences fully participate in nonhost resistance [3]. Such defences can take place at the anatomical (cuticle or cell wall appositions) or at the biochemical level (secondary metabolism and antimicrobial proteins or peptides, the plant defensins [4,5]). Nonhost resistance may also result from the induction of defence responses activated by the plant's ability to detect the presence of exogenous eliciting molecules like fungal elicitins [6] or bacterial flagellin [7]. In this particular case, the induced resistance is linked to the establishment of a much localized necrotic response occurring at the infection site, the hypersensitive response (HR).
2.2 Race-cultivar specificity
This level of specificity occurs during a host relationship when a genotype of an otherwise susceptible plant species exhibits resistance to a pathogen genotype. It involves:
- – cultivars (cv.) or varieties of plants;
- – races of fungi or bacteria;
- – strains of viruses;
- – races, biotypes, pathotypes or strains of nematodes.
The parasite pathogenicity is then expressed by the term ‘virulence’ sensu stricto and its lack of pathogenicity by the term ‘avirulence’. The plant is the place where either a compatible (parasite virulence and host plant susceptibility) or an incompatible (parasite avirulence and host plant resistance) reaction is expressed. The corresponding resistance has been referred to as ‘vertical resistance’ by van der Planck [8]. Its expression is independent of environmental conditions and vertical resistances are most often mono- or oligogenic [9].
Two types of mechanisms involved in race-cultivar specificity have been described so far. First, the ‘specific toxin’ system that involves the production of cultivar-specific toxins by the pathogen. This system is exemplified by resistances to necrotrophic fungi and the Hm1 gene from maize cultivar resistant to race 1 of Helminthosporium carbonum, which was the first race-cultivar specificity-encoding gene to be cloned [10]. Secondly, this type of specificity is ruled out by the famous gene-for-gene relationship and the associated Flor's model that established a tight genetic link between the host and the parasite [11]. It was after Flor's work that any gene conferring a vertical resistance in race-cultivar specificity was called a ‘resistance gene’ or ‘R gene’. Flor [12] showed that the resistance of a flax cultivar to a Melampsora lini rust race was conditioned by the simultaneous occurrence of a plant R gene and a pathogen avirulence gene (Avr gene). These two genes were shown to be dominant and the interaction turned out to be incompatible when the two genotypes possessed both genes corresponding to each other. In other situations, i.e. with the occurrence of r and/or avr recessive alleles, the interaction was compatible.
Many R genes have been isolated and characterized during the last decade. The first cloned R gene was Pto, which confers resistance to tomato against the bacterial pathogen Pseudomonas syringae pv. tomato [13]. Gene tagging and map-based cloning were the most successful and extensively used techniques to clone these genes [14], from plants of great agronomic importance such as cereals (wheat, barley, maize and rice) and several dicots (tomato, tobacco, flax, sugar beet, potato and lettuce) but also for the small model weed Arabidopsis thaliana. Moreover, the extent of cloned R genes corresponds to resistances against most pathogenic organisms, from viruses to nematodes [15,16]. The comparison of the predicted structure of R genes products quickly revealed that common structural features were shared among them, with each structural domain (leucine reach repeat, nucleotide binding site, coiled coil, ...) exhibiting a putative function in the expression of resistance at the cellular level [16].
Moreover, loci corresponding to R genes have also been extensively studied. In these genetic studies, any locus conferring race-specific resistance on a plant cultivar is called ‘specificity’. These specificities have been shown to exhibit a particular distribution within the plant genome: many clusters of R loci encoding for several specificities have been found, especially in the case of resistances to biotrophic fungi [11]. Two types of genomic organization accounted for this non-random distribution: allelic variation and chromosomal tandems and clusters of tightly linked genes [15,17]. This distribution of R loci in the plant genome identified two major issues. These issues are the origin of this organization and distribution, and the evolution of R loci in connection with diversity generation and maintenance [17–19], both of which being currently extensively studied.
2.3 The plural nature of resistance in plants
One has to emphasize the fact that the specificity levels in plant–pathogen interactions are not limited to those mentioned above. For instance, tissue and organ specificities are now receiving more interest than in the past and it can be expected that parasitic factors conditioning the ability to invade a given tissue or organ will be characterized in the near future [20,21]. Moreover, the elegance of the Flor's model and the gene-for-gene relationship, together with the tremendous progresses made in the understanding of corresponding molecular events, may suggest that the HR taking place during race-cultivar specificity and the interaction between the products of Avr and R genes is, if not universal, the predominant mode of expression of resistance to pathogens in plants. The increasing number of publications concerning this particular type of resistance is probably responsible for the heightened awareness of this mechanism. However, one has to keep in mind that large differences in the expression of resistance certainly reflect an important diversity in parasitic strategies. Far from being an exhaustive list, the following are some of the most common types of resistance which can be observed in nature or mentioned in the literature.
An ‘innate’ resistance (which is distinct from ‘innate immunity’, see conclusion below) can potentially be expressed by plants prior to any contact between them and their pathogens. Preformed defences, nonhost resistance and incompatible reaction during a gene-for-gene relationship that have been mentioned above belong to the resistances that plants possess per se. On the other hand, ‘acquired’ resistances are expressed only by plants on which a conditional potential for resistance has been conferred. Two types of acquired resistances are widely cited in the literature: the local acquired resistance (LAR), expressed in a localized manner and restricted to the site of resistance acquisition, and the extensively studied systemic acquired resistance (SAR). For the latter, in many interactions, the establishment of HR results in a non-specific and systemic resistance to a wide range of pathogens [22]. This type of acquired immunity can last for several weeks.
In plants, the word ‘acquired’ in ‘acquired immunity’ is a good example of terminological ambiguity. Indeed, it is a clearly distinct concept from acquired immunity in animals, which is highly specific and in contrast to what is often referred to as ‘acquired resistance’ in plants. Finally, induced resistance (IR) is expressed after a biotic or non-biotic treatment, which makes a plant less susceptible, tolerant or even fully resistant. Both ‘induced’ and ‘acquired’ resistances in plants are terms often used interchangeably with the same meaning.
2.4 Plant and pathogen factors involved in plant–pathogen interactions
Beside specificity levels, basic compatibility between a given plant and a given pathogen results from the balance between plant resistance factors – the ‘general’ resistance factors – and the pathogenicity determinants of the pathogen [2]. Pathogenicity is then referred to as either ‘virulence’ sensu lato or ‘aggressiveness’. The plant resistance can be represented in a quantitative manner as a continuous parameter. Quantitative resistances, which have been termed ‘horizontal’ resistances according to van der Planck's terminology [8], are often polygenic and environmental factors exert such an important influence on the outcome of the interactions in which they are involved that the environment can be considered as a third partner in the interaction [2].
For basic compatibility, mechanisms of general defences have to be considered. These are the active mechanisms induced by the recognition of:
- (1) a pathogen by the plant,
- (2) the molecular pattern associated with it (see below in the conclusion), and
- (3) an elicitor released from this pathogen (exogenous elicitor), or
- (4) an elicitor released from the plant attacked by the pathogen (endogenous elicitor).
Now consider the issues of basic compatibility from the vantage point of the parasite and the defining of factors enabling pathogens to colonize the host plant. The parasite must overcome many significant impediments in order to invade the host, induce symptoms and still complete its life cycle. The obvious temptation would be to distinguish between the general biological ‘house-keeping’ function and those that are involved in pathogenesis only [34]. It could therefore be postulated that such a factor would be intrinsically involved in pathogenicity in natural conditions and is unnecessary for vital functions. However, the case has to be considered of obligately biotrophic fungi, for which house-keeping and pathogenicity functions are much more difficult to distinguish than for facultative parasites. Moreover, a more complete point of view is particularly necessary when pathogenic factors that make the plant defence inefficient are considered [35]. Eventually, a global approach of pathogenicity was proposed for animal pathogens, but may also be useful for plant pathologists: a pathogenicity determinant is a microbial product that influences the progression of the infection [36]. In the case of necrogenic bacteria, that eventually kill the invaded plant tissues, the most important pathogenicity factors to be considered are exopolysaccharides of surface (EPS) [37], hydrolytic cell wall degrading enzymes [38], toxins [39], or other diverse factors such as iron acquisition systems [40], and the synthesis of auxin-like growth regulators [41]. These bacteria also use intercellular signals involved in quorum sensing that activate the expression of numerous genes during infection [42]. Concerning filamentous fungi (fungi sensu stricto or filamentous parasitic protoctista such as Oomycetes), three strategies are currently used in order to determine which genes are encoding for pathogenicity determinants. These strategies are:
- (1) gene disruption or replacement of a gene encoding for a known function a priori important in pathogenicity [34,43],
- (2) random mutagenesis techniques such as gene tagging, differential display, sequencing of ESTs derived from transcripts accumulating during infection [44], macro and micro-arrays investigations, and
- (3) the application to plant pathogenic fungi of the functional genomics technologies that allow insertional mutagenesis at the scale of the entire genome.
Currently, the principal pathogenicity factors that have been identified so far are linked to the early stages of infection (spore adhesion and germination, plant surface recognition and differentiation of infectious structures like appressoria), the production of toxins, the release of cuticle and cell-wall degrading enzymes, and the detoxification of plant antimicrobial compounds [34,46].
A model representing the different components of interactions as they are classically described is presented in Fig. 1. The central theme of this review will focus on the fact that distinctions included in this model can now be seriously questioned.
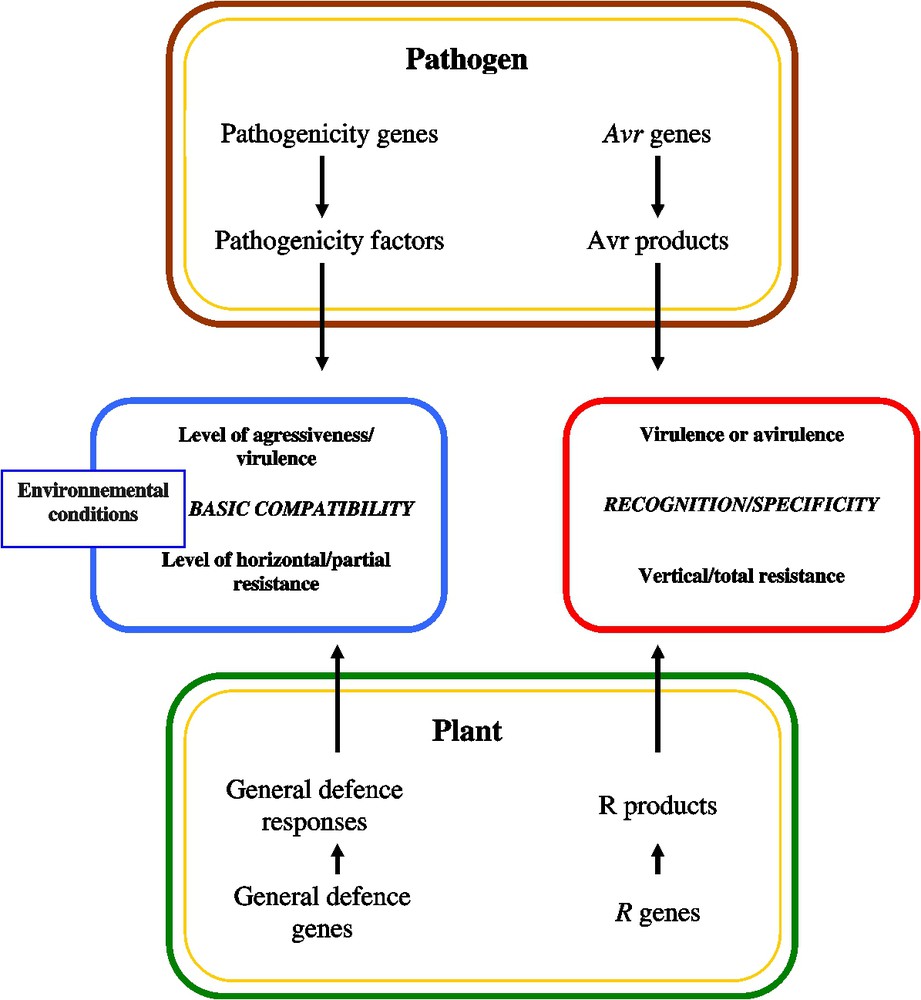
A classical model for the plant and parasite components involved in phytopathogen interactions.
3 Is it still accurate to consider separately pathogenicity and resistance?
First of all, a direct link between bacterial aggressiveness, as a component of pathogenicity in basic compatibility, and avirulence, which determines the plant resistance when race-cultivar specificity occurs, appeared with the discovery of the hrp (hypersensitive response and pathogenicity) system in several species of necrogenous bacteria. In Pseudomonas syringae, Ralstonia solanacearum, Xanthomonas campestris and Erwinia amylovora, studies involving random mutagenesis allowed the isolation of hrp mutants. These mutants showed altered pathogenicity on susceptible plants and they lost their ability to induce the HR during nonhost or cultivar-specific resistance. It has been shown that more than 20 hrp genes are clustered in about 20 kb in the bacterial genome [42]. These genes encode for proteins of the type-III secretion system, which allows the direct delivery of effector proteins probably involved in pathogenicity: HrpN in Erwinia amylovora, HrpZ in Pseudomonas syringae or PopA in Ralstonia solanacearum [47]. The precise role of these proteins remains to be investigated, although hrp− mutants of Erwinia chrysanthemi are clearly altered in pathogenicity. Some possess LRR domains like most of the R products, and they may become anchored to the plasma membrane, incorporated into the nucleus or participate in the formation of bacterial pili [48]. The avirulent phenotype of the hrp− mutants is due to the fact that Avr products are secreted via the same type-III system as these proteins [42].
Race-cultivar specificity for phytopathogenic fungi also involves factors classically described only as basic compatibility factors. Xylanase activity is a typical example of the basic compatibility factor produced by Trichoderma and has been shown to induce a monogenic resistance in particular in tobacco cultivars [49]. This resistance is therefore very close to a classic vertical resistance. Genes encoding for receptors to xylanases inducing such a resistance have been cloned recently from tobacco and tomato. Predicted products of these genes have been shown to share the same structural features as products from previously cloned and characterized R genes [50]. The xylanase protein is therefore now to be considered as a determinant of both pathogenicity and aggressiveness and as an Avr product.
Another link between the expression of plant resistance and the expression of pathogen aggressiveness has been established with the characterization of the HrpW harpin protein from Pseudomonas syringae and Erwinia amylovora, which is responsible for the expression of an incompatible reaction in the attacked plant. The HrpW protein contains both an N-terminus homologous to the other harpins and a C-terminus homologous to a novel class of fungal and bacterial pectinases [51]. The cloning of the Cf-9 gene in tomato revealed a strong homology with genes encoding plant polygalacturonase-inhibiting proteins (PGIPs) [52]. Since then, PGIPs have been shown to have other similarities with either extra- or intracellular R gene products and with a flagellin receptor [53].
The links between basic compatibility and resistance were recently strengthened by the study of Arabidopsis thaliana mutants, which are unable to develop their preformed constitutive defences, but are still able to perform the HR when challenged with avirulent strains of biotrophic fungi. They are also altered in their compatibility with virulent strains [54]. Such a reported plant compatibility factor is encoded by the PMR6 gene, which showed a strong homology with a pectinase-encoding gene [55].
Since cellular and molecular links between the pathogenicity determinants and vertical or horizontal, induced or preformed plant resistances are now found more often in the literature, it now appears clear that the concepts of both pathogenicity and resistance are mutually necessary for each one to be understood.
4 To what extent can we distinguish between qualitative and quantitative resistances?
As predicted by Nelson [56] and Crute [9], the distinction established between quantitative and qualitative aspects of plant resistance is not always discernible. According to Nelson [56], quantitative resistances are the result of residual cumulated qualitative resistances. For Crute [9], if a compatible interaction is necessarily conditioned by a mutual recognition of both partners, the severity of the resulting symptoms is clearly determined by a more basic compatibility. It was justified onwards to address the question of whether the different types of plant resistances and the different types of interactions they establish with their parasites are parts of a complex continuum. The occurrence of this continuum seems to be at least partially verified today when cellular and molecular mechanisms are taken into account in the study of these concepts. Recent studies on Arabidopsis thaliana clearly illustrate this hypothesis: the quantitatively continuous range of phenotypes obtained on different plant accessions challenged with different Peronospora parasitica isolates is not consistent with the either totally incompatible or totally compatible reactions that are supposed to characterize the gene-for-gene relationship [57]. With regard to defence responses, it is well known that vertical and horizontal resistances involve a common set of defence mechanisms, although these defences are distinct in their kinetics and intensity [58].
Accordingly, a vertical resistance involves general resistance factors classically associated with horizontal resistance: tomato cultivars resistant to several races of Alternaria solani have been shown to exhibit a very important constitutive transcription level for several PR-proteins encoding genes [59]. The genetically engineered overexpression of the Pto gene conditioning race-cultivar specificity between tomato and the expression of the HR leads to the activation of general defence responses and to a resistance extended to other bacteria and also to fungi [60]. Recently, the ‘guard hypothesis’ has been proposed as a model for the mechanisms of the Avr–R interaction and recognition and is also an additional illustration of these links between qualitative and quantitative aspects of interactions. According to the ‘guard hypothesis’, the Avr product, which can in some cases be a pathogenicity determinant ‘recognized’ or ‘sensed’ by the plant, interacts with a host plant protein, the so-called ‘pathogenicity target’. The most described and characterized pathogenicity target, the RIN4 protein from Arabidopsis thaliana, is associated with suppressing activity of the host's general defences; this suppression is reinforced when an interaction with an Avr product occurs. It appears that the corresponding R product is able to ‘guard’ against or ‘sense’ for the presence of the molecular complex formed by the Avr product and the pathogenicity target. This mechanism prevents the general defence responses inactivation and induces the HR [61,62]. Besides the common mechanisms shared by the general defences of quantitative resistances and the Avr recognition of qualitative resistances, the ‘guard hypothesis’ also illustrates the links between resistance and pathogenicity that have been mentioned above.
This continuity between qualitative and quantitative resistances may result from distinct but overlapping features of the defence responses range that would be involved in both cases [3]. At the genetic level, there is growing evidence of the overlap of these two types of resistance. This is illustrated by the study of quantitative trait loci (QTLs) leading to the resistance of the French bean to Colletotrichum lindemuthianum. QTLs responsible for this partial resistance have been co-localized in the bean genome with R genes, but also exhibited a clear specificity to different isolates of the fungus [63]. The reports of such a co-localization in the genome of QTLs and R genes are also found more frequently in the literature [64]. Moreover, novel R or defence response-encoding genes have been sought based upon the search for and the mapping of resistance gene-like sequences (RGL) or resistance gene analogues (RGA) from rice, maize and barley. This led to evidence that these sequences were tightly linked in the rice genome to both quantitative and qualitative resistance loci [65]. About ten years ago the strong genetic linkage between a resistance QTL to the Oomycete Phytophthora infestans, in potatoes, which correlates with late maturity and the race specific R1 gene led to the hypothesis that both types of resistance had molecular links [66]. Subsequently, it has been shown that the R1 gene belongs to the NBS-LRR class of the R gene family [67] and that the resistance QTL also co-localizes with PR-proteins encoding genes [68]. RGL sequences have also been co-localized with other resistances in the potato genome [69]. PR-proteins encoding genes and RGL sequences are now true candidates for QTL genes for partial resistance to P. infestans [70]. Finally, race-specific QTLs have been also reported [71], for example in the case of partial resistance of apple tree to Venturia inequalis [72].
5 Is there a single type of resistance in plants?
Other differences in the types of resistance – host and race-cultivar specificities, compatible and incompatible interactions, preformed and induced defences – do not necessarily hold true when subjected to molecular and cellular investigations of the interactions. In many recent investigations, the borderline between compatible and incompatible interactions has been shown to be indistinguishable, at least at the experimental level. When syringolin A, a bacterial determinant for nonhost resistance, is applied to wheat plants involved in a compatible interaction with Blumeria graminis f.sp. tritici, it induces a conversion of the interaction into an incompatible one, leading to a typical HR [73].
Moreover, two main hypotheses have been already proposed to explain the links between host and race-cultivar specificities [74]:
- – nonhost resistances, as well as qualitative resistances (see above), result from the cumulated action of many R genes, each of them encoding for its own specificity, and would therefore be determined by a range of recognition events between R products and elicitors/Avr products;
- – alternatively, nonhost resistances involve a unique gene-for-gene relationship similar to that of race-cultivar specificity.
Many recent works clearly show that common mechanisms govern these two previously distinct specificities, which can now be classified as a single phenomenon. It has been shown that both plant resistance-encoding genes – R genes and others – and microbial elicitors could be involved in these mechanisms. The RPW8 gene from Arabidopsis thaliana was initially characterized as a classic gene-for-gene relationship between a plant ecotype and the corresponding race of Erysiphe cichoracearum. It then turned out to encode for resistance in at least 18 races among four different Erysiphe species and then to all other tested races [75]. The RPW8-encoded resistance therefore clearly tends towards host specificity. In marigold, a gene encoding for a TIR domain that is commonly found in many race-specific R products is induced during nonhost resistance to the parasitic Phanerogam Striga asiatica [76]. The non-specific bacterial elicitor flagellin [77] is recognized in Arabidopsis by a receptor with a kinase activity and LRRs that are also found in many race-specific R products [78]. In tomato, the HR-associated recognition of fungal proteins secreted by Cladosporium fulvum occurs in both host and nonhost plants [79]. In wheat, nonhost resistance to Blumeria graminis f.sp. secalis, responsible for rye powdery mildew, involves a gene-for-gene relationship between plant genes and fungal genes encoding for a non-specific avirulence [80]. In pea, non-specific and nonhost resistance involve the specific recognition of the elicitor encoded by the AvrPphD gene of Pseudomonas syringae pv. phaseolicola [81].
Cellular events and transduction pathways induced in plants either by an R-Avr recognition event or the binding of a non-specific elicitor also share many common features. A study of transduction pathways activated by R genes from the three different classes, the RPW8 gene or by a compatible interaction showed that the same pattern of genes and the synthesis of PR-proteins are also induced [82]. In Nicotiana benthamiana, the SGT1 protein illustrates the occurrence of such common effectors involved in both nonhost and race-cultivar resistances [83]. As already mentioned above, at the cellular level, HR has been shown not to be restricted to the expression of incompatible reactions when race-cultivar specificity occurs. For instance, HR was shown to be associated with both nonhost and race-cultivar resistances of Solanum species to the Oomycete Phytophthora [84], suggesting that identical mechanisms were involved in both cases. HR is observed in many different contexts, from the nonhost resistance to necrotrophic pathogens to race-cultivar resistance to viral genomes. In addition to the formerly known elicitins (see above) a growing number of HR-inducing proteins have been revealed [85] outside of the initial race-cultivar background. Cell wall reinforcement such as papilla deposition is another example of common defence mechanisms involved in both types of resistance [86]. The importance of intracellular vesicular trafficking in papilla deposition during host and race-cultivar specificities in barley and Arabidopsis has been recently shown [87]. However, it must be mentioned that even when they induce HR, plant cells involved in host or nonhost resistance do respond in different ways: in Vigna and French bean, nonhost HR appears earlier and involves intervacuolar strands, Brownian movements and nucleus migration to the penetration site. It may also be distinguished from host HR using pharmacological inhibitors [88]. At the genomic scale, the comparison of Arabidopsis transcriptomes corresponding to either host or nonhost resistance to the bacterium Pseudomonas syringae shows that one fourth of the expressed genes are regulated the same way in both cases [89].
Finally, other close links have also been found between preformed and induced defences, in addition to those mentioned above in relation to plant compatibility factors. Potato PR-proteins encoding genes showed unexpectedly high expression levels and are suspected to be involved in non-specific partial and constitutive resistance to P. infectans [90]. Moreover, the challenging, by a tomatinase-deficient strain of Septoria lycopersici, of Nicotiana benthamiana, in which defence response genes had been silenced, revealed how the pathogen was able to bypass the plant defences, using a two-step process: the hydrolysis of the antimicrobial plant saponin, tomatin, which is considered a classic preformed plant defence, and the suppression of induced defence responses by the released product of the hydrolysis [91].
6 Conclusion: to what extent will the concepts be unified?
It is now well established that all plant pests, including pathogens and herbivore insects, induce a modification in the expression of plant genes via the recognition of eliciting molecules as well as at least an alteration of cell and tissue integrity. Many recent molecular and genomic studies of plant responses to biotic and non-biotic stresses suggest strong connections inside the plant cell between transduction pathways allowing both the animal and microbial ‘non-self’ to be recognized, as well as responses to environmental stresses [92]. A new updated model for interactions between plants and pests has been proposed and is presented in Fig. 2.
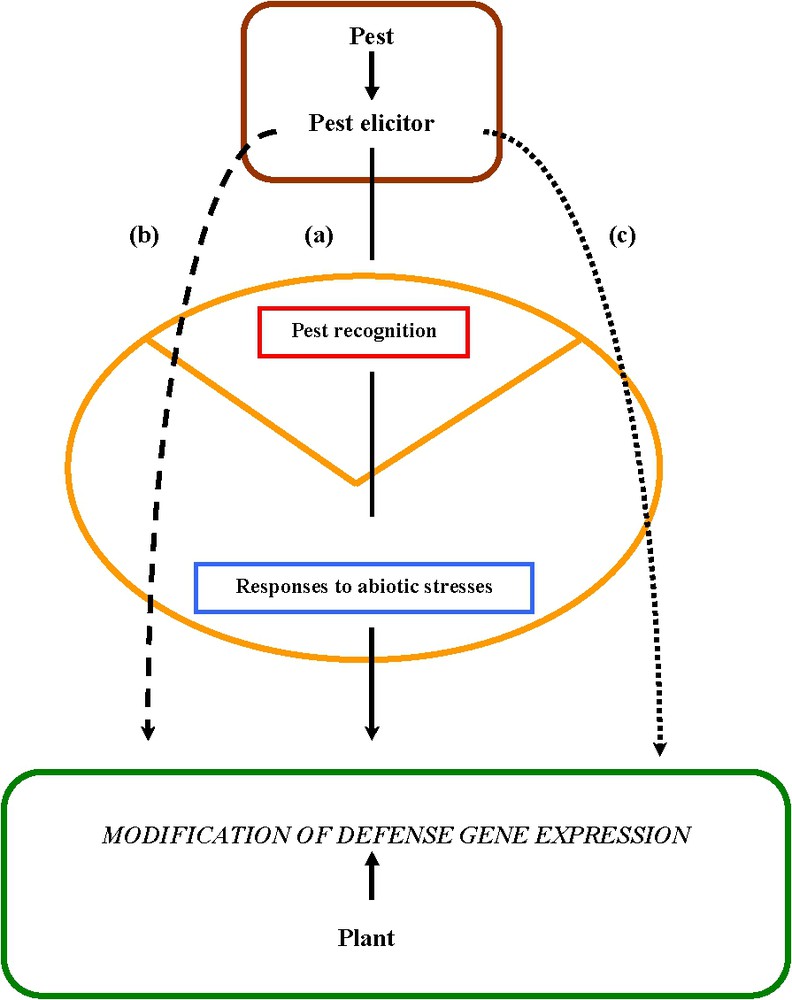
An updated and unified model for plant–pest interactions (according to [92]). Transduction pathways activated by the recognition of a parasite or an herbivorous insect are connected together, as well as with pathways involved during the response to abiotic stresses. The expression of plant genes involved in these transduction pathways is modified whatever the extent of the pest recognition by the plant. (a) The pest elicitors are fully recognized by the plant. (b) Modified elicitors are produced and only partially induced the defence transduction pathway. (c) Modified elicitors are produced, they are not recognized by the plant and do not induce the defence transduction pathways.
The homogenization of the concepts related to plants might logically be extended to animal pathology concepts as well. Nevertheless, several fundamental differences seem to make mechanisms of plant and animal pathologies irreconcilable: the lack of plant mobility, the absence, in plants, of an immune system involving specialized and circulating cells, the presence in mammals of the major histocompatibility complex (MHC) leading to the recognition of non-self by specific antibodies and the fact that, in plants, all cells have the same genetic defensive potential.
However, concerning pathogenicity, it has been shown with type-III secretion [47] and iron acquisition [40] systems that animal and plant pathogenic bacteria share common features. In Pseudomonas aeruginosa, clusters of genes encoding for pathogenicity in animals found together with genes encoding for pathogenicity in plants have been identified [93]. Finally, the Pseudomonas YopT and Xanthomonas AvrBsT proteinous effectors showed the same cysteine proteinase activity as YopJ from the animal pathogen Yersinia. This activity has been associated with alterations of the host cells [94,95].
Concerning resistance, defensins are a clear example of antimicrobial proteins produced by both animal and plant organisms [96]. Moreover, the complex non-random organization of R loci clusters and tandems as well as the evolution mechanisms of these loci is now considered to be similar to the HMC in mammals [97].
A new model has been recently proposed [17] to explain the proliferation of R genes and their complex organization within plant genomes. This model relies on the principle of the HMC and the functioning of immunoglobulin-encoding genes [15]. According to this so-called ‘birth-and-death’ model, complex R loci would be originated from initial duplication events. Indeed, transposed sequences have been found within complex R loci that probably caused the duplication events. The number of R genes would then have increased because of unequal cross-over events between mispaired gene copies during meiosis. The molecular location of these events is likely to be conserved leucine reach repeats (LRRs) between copies. They can take place either between genes, causing the number of gene copies to vary, or within a given gene, making the number of LRRs vary. Such recombination events along with genic conversions or transposon insertions would create some sequence divergence between the generated gene copies resulting in the so-called ‘death’ of the locus. Indeed it has been shown that paralogue genes, which are located within the same cluster, are more divergent from each other than orthologue genes, which are alleles in haploid genomes [19]. However, two major issues remain to be investigated, which despite extensive molecular investigations and the recent development of the ‘birth-and-death’ model have not yet been addressed:
- – what is the nature of the events originating the selection pressure leading to the sequence divergence?
- – why are R loci organized either in tandems of clustered genes or in allelic series?
In Arabidopsis thaliana, RPP genes encoding specificities to the Oomycete Peronospora parasitica are a good model for the study of R genes polymorphism in natural plant ecotypes and they proved to be useful in the understanding of polymorphism preservation among them. Two hypotheses have been suggested [15]:
- – when overcome, and therefore ineffective, because of the emergence of a new virulent race of the pathogen, R genes disappear from the plant population. They only persist during a temporary period, which is determined by their efficiency against the pathogen population;
- – alternatively, such overcome R genes are maintained at a low frequency within the population, until the corresponding avirulent race of the pathogen does appear again within the pathogen population. In this case, they are recycled R genes. This second hypothesis is the most widely accepted today: many cloned R genes have been shown to possess alternative alleles within accessions distinct from those from which they have been isolated. A certain amount of polymorphism is therefore generated, accumulated and maintained in this way between different accessions.
Since RPP5, N and genes have been cloned, it is known that intracellular transduction pathways leading to resistance are sometimes highly conserved between organisms, even in the unexpected case of plants and drosophila, for example. The strong homology of the TIR domain from many plant R genes with the interleukin receptor shows such a link between animal immunity and plant resistance. This TIR domain is also homologous to the drosophila Toll receptor which is involved in defensin production and innate immunity [97], as well as in embryogenesis [98]. A Toll homologue has been characterized in mammals and shown to also be involved in innate immunity [99]. Moreover, innate immunity in mammals also involves NOD intracellular receptors exhibiting strong homologies with NBS and LRR domains from the plant R genes [97]. Plant and animal resistances to disease therefore involve a growing number of similar mechanisms, like plant HR and animal apoptosis, but also conserved transduction pathways and identical structural domains [100,101].
Surprisingly, although the accumulation of cellular and molecular work renders classic concepts and associated distinctions at least partially unified, new and additional concepts are also emerging. As an example, the concept of a general elicitor may be on the way to being replaced by the term ‘pathogen-associated molecular pattern’ (PAMP), which has recently been imported from animal pathology. As with PAMPs recognized during animal innate immunity, the pep-13 elicitor from Phytophthora infestans shows a structure highly conserved between different Phytophthora species and the bacterial flagellin is also found in several bacterial plant pathogens. Both are recognized in plants and have been shown to elicit defence responses [102]. Is the emergence of PAMPs in plant pathology just another step towards the unification of plant and animal pathology concepts rather than the emergence of a new concept in phytopathology? New distinctions can also emerge: acquired resistances like SAR are now separated from induced resistance such as the induced systemic resistance (ISR) [103]. SAR is now mentioned as a SA-dependent mechanism, whereas ISR is reported as SA-independent, but involving jasmonic acid and ethylene. ‘Induced accessibility’ and ‘induced inaccessibility’ are also mentioned in a growing number of recent works. A plant cell challenged with a compatible pathogen becomes accessible to a previously incompatible one. General, non-specific, race-specific and nonhost resistances have been shown to be altered in this way. On the other hand, an otherwise susceptible cell is rendered inaccessible to a compatible pathogen when it has been previously challenged with an avirulent one [54]. Another new ‘enzymatic resistance’ concept has also been proposed recently [104]. This resistance has been associated with the constitutive expression of genes involved in normal non-pathogenic metabolism, encoding for factors regulating carbon metabolism or for different enzymatic activities such as adenosine kinase, glycerol kinase or activities involved in photo-respiration.
However, it remains clear that the experimental progress over the past 15 years, and especially that of the past five years, strongly encourages phytopathogenic interactions to be conceived in a more global fashion than they once were classically. The molecular and cellular dissection of mechanism underlying these interactions indicate that the limits and separations between the different concepts mentioned above are not as much justified as they used to be. But is this new global vision resulting from studies performed at the micro- or the nanoscale still relevant in the field? Will the different resistance types stated by van der Planck [8] and the other classical concepts not be relevant anymore for studies of resistance management in the field or for epidemiological investigations? The answer to this question will depend upon how successfully molecular and cellular investigative techniques can be implemented in actual large scale, real time studies and investigations. New defence strategies based on molecular breeding and the use of GMOs are often prohibited in Europe based on social and economic grounds, but also because of the agronomic cost of transgene expression and its consequent effects on yield and plant growth. Provided these obstacles are overcome at some point in the future, it should be possible to verify whether the field, as a laboratory, becomes a proper stage for the unification of the concepts linked to plant–pathogen interactions.
Acknowledgements
We are grateful to Shannon Neil, Clay Goldberg and Dr. D. Lösel for their comments on the manuscript and their help in revising the English text.


