1 Introduction
All metazoan phyla contain organisms that undergo adult regeneration. Simple organisms, such as the cnidarian hydra polyp or the ‘primitive’ bilaterian planaria, are able to regenerate their main body axes in both directions, while more complex metazoans, such as salamanders, regenerate large, multi-tissue structures such as limbs, tails, jaws, and tissues of the eye. In these animals, the developmental programs remain accessible to reactivation, whatever their age. This widespread distribution suggests that regeneration is an ancient feature of multi-cellular organisms that was maintained in some organisms and lost during evolution in others [1]. In fact, in mammalian species, this regenerative potential is progressively lost during development, although tissue regeneration remains possible, like that of the skin and its ectodermal derivatives, the bone, the liver, the lens, and partially the nervous system. This seemingly random distribution of regenerative capacities among the Metazoa is puzzling, and little is known about the genes and genetic programs responsible for this biological trait.
The freshwater hydra polyp is a classical model system to investigate the cellular and molecular basis of regeneration, as hydra polyps exhibit amazing budding and regenerative capacities whatever their age. Hydra displays a tube shape with a single opening at the top, circled by a ring of tentacles, which has a mouth-anus function, whereas the basal disk attaches to the substratum (Fig. 1). Like all cnidarian polyps, hydra is formed of two cell layers, with differentiated tissues and/or structures at the extremities but no organs as recognized in bilaterians. The ectodermal myoepithelial cell lineage provides the outer cell layer, while the endodermal myoepithelial cells together with the gland cells line up the gastric cavity. A third distinct pool of cells, the interstitial cells, comprises common stem cells for the somatic and germinal cell lineages, including neurons, mechanoreceptor cells (nematocytes), gland cells, and gametes. Surprisingly, hydra can regenerate their head in the absence of cell proliferation as well as in the absence of neurogenesis. These observations led to the assumption that hydra regeneration occurs through cellular processes that are distinct from those characterized in bilaterian species, as planarians and urodeles (see in [2]).
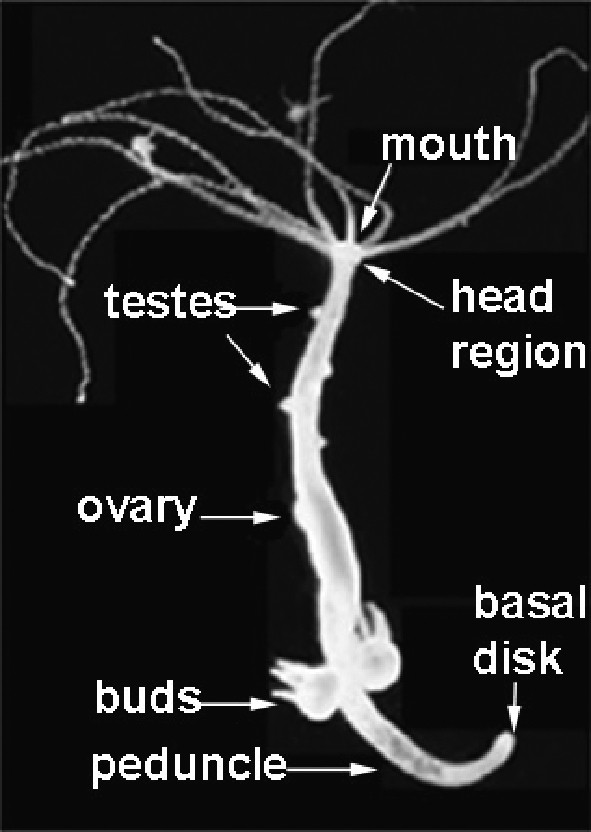
Hydra is a freshwater hydrozoan that lives as a polyp exclusively. Anatomy of the hydra polyp: the head region, the body column, the budding zone, and the basal disk. Testes, shaping as nipples, can be seen in the upper half of the body column.
The discovery of the conservation of the homeobox motif between Drosophila and vertebrates genes as well as their chromosomal organization and functional hierarchy proved that pieces of evolutionarily conserved developmental pathways were recruited for similar developmental tasks by protostomes and deuterostomes [3]. Such functional conservation between bilaterians implies that these developmental pathways were already present in their common ancestor, named ‘urbilateria’ [4]. In the absence of currently living ‘urbilateria’ species, cnidarians and ctenophores are the best candidates for investigating a non-bilaterian representation of these ancestral developmental pathways.
The hydra regulatory genes actually proved to be highly conserved [5–7], and their expression patterns analyzed at the developmental and cellular levels provided a first step towards a dynamic view of their regulation. In fact, the comparison of the expression patterns observed in the adult polyps with those detected in budding, regenerating, or re-aggregating animals implies that the genetic networks involved in the cellular reprogramming at the time the animal initiates regeneration are likely differently regulated from those maintaining homeostasis in the adult animals. At the temporal level, distinct sets of genes are regulated after amputation, either immediately (within 1 h), or early (within 10 h), or early–late (from 15 to 36 h) or late (after 40 h). The ‘immediate’, ‘early’, ‘early–late’ waves of gene regulation arise concomitantly with the wound-healing phase, the establishment of organizer activity and the differentiation of head structures respectively, the ‘late’ genes being re-expressed in the newly differentiated head [2,6,8]. In addition, most of the early-induced transcripts are restricted to the endodermal cells of the regenerating tip.
To obtain gene-specific loss-of-function assays, we applied to hydra the procedure previously developed for nematode and adapted to planaria, i.e. feeding the animals with bacteria producing dsRNAs [9,10]. This method leads to an efficient, harmless and incremental RNAi silencing [11], and produced a variety of distinct phenotypes in regenerating hydra ([11,12], SC, unpublished), opening hence the possibility to decipher the genetic control of the cellular remodelling underlying head regeneration.
2 Three major phases in head regeneration
2.1 The immediate phase: the serine protein-kinase inhibitor Kazal1 as an essential cytoprotective agent
In the freshwater hydra, the gastrodermis is made up of gland cells that produce zymogens and express Kazal1, a serine protein-kinase inhibitor Kazal-type (SPINK) gene, as well as endodermal epithelial cells that behave as digestive cells. The Kazal1 function was tested in hydra fed with bacteria expressing dsRNAs. Those showed a progressive decrease in Kazal1(–) transcript abundance, decrease that affected homeostatic conditions as evidenced by the low budding rate and the induced animal death, already detected after three feedings. Concomitantly, a dramatic disorganization of gland cells followed by their massive death was observed, while their neighbouring cells, the digestive cells, displayed a highly vacuolated cytoplasm. Those vacuoles were assigned as autophagosomes as they contained mitochondria and late endosomes.
Amputation of wild-type (wt) hydra leads to an enhanced Kazal1 expression in regenerating tips (Fig. 2, left panel). However, in Kazal1 knocked-down hydra (Fig. 2, right panel), autophagosomes were immediately detected in endodermal cells of the regenerating tips, and upon complete silencing, hydra no longer survived the amputation stress [11]. This first cellular phenotype resulting from a gene knockdown in cnidarians highlights the essential digestive and cytoprotective functions played by the Kazal1 serine-protease inhibitor activity in hydra. In mammals, autophagy of exocrine pancreatic cells is also induced upon SPINK1/SPINK3 inactivation, whereas SPINK3 expression is activated in injured pancreatic cells [13–15]. Hence, SPINKs, by preventing an excessive autophagy, appear as key players of the stress-induced self-preservation program. Enhancing the self-preservation program in injured tissues might therefore be the condition for unmasking their potential cell and/or developmental plasticity [16].
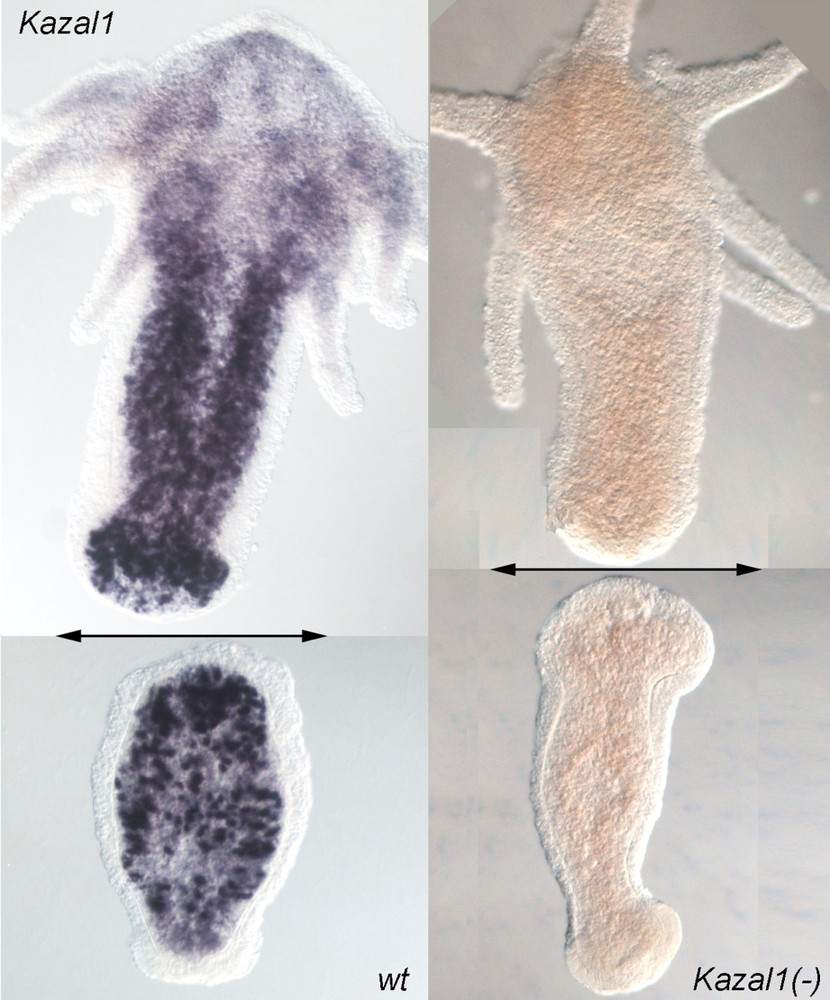
Genetic function in hydra inferred from RNAi silencing. Kazal1 expression in regenerating hydra 4 h after bisection. Bisection level is indicated with a double arrow. Upper halves regenerate their foot in two days, whereas lower halves regenerate their head in three days. The massive Kazal1 expression in gland cells (left) is required for protecting the cells against the amputation-induced autophagy. In Kazal1(–) hydra, obtained after repeated dsRNAs feedings (right), hydra no longer survive the amputation stress.
2.2 The early phase is under the control of the RSK / CREB pathway
Following a distinct strategy, we used DNA-binding assays to screen for transcription factors that would exhibit modulations in their DNA-binding complexes after amputation. That way, we identified and cloned the cAMP Response Element Binding (CREB) protein, which shows three highly conserved domains: the kinase inducible domain (KID), the DNA-binding and dimerization leucine-zipper domains (bZIP) [17]. In vertebrates, the CREB transcription factor mediates the response to a large array of extra-cellular signals to the nucleus through post-translational modifications that involve multiple protein kinases [18], including the RSK kinase [19]. This kinase phosphorylates a particular residue, Ser133, located in the KID, an event that is critical for modulating CREB transcriptional activity, namely because the phosphorylated form of CREB specifically binds to the ubiquitous and multifunctional transcriptional co-activator CBP [20].
In hydra CREB, the Ser67 residue located in KID is a target for post-translational regulation, similarly to the Ser133 residue characterized in the CREB vertebrate proteins. By using the anti-phosphoSer133-CREB antibody together with the antihyCREB antibody, we noted that phosphoCREB-expressing nuclei were restricted to the endodermal layer, while CREB-expressing nuclei were distributed in both layers. Interestingly, immediately after amputation, the number of phosphoCREB-expressing nuclei increased significantly in the head-regenerating tips (Fig. 3A). Biochemical and immunological evidences identified a RSK-like kinase, which showed an enhanced activity and a hyperphosphorylated status during head but not foot regeneration [21]. Exposure to the U0126 MEK inhibitor, which prevents RSK phosphorylation, inhibited head but not foot regeneration (Fig. 3B), while in head regenerating tips, CREB phosphorylation was abolished and the early gene HyBra1 was not activated. These data support a role for the MAPK/RSK/CREB pathway in one specific cell lineage, the endodermal myoepithelial cells, likely linked to the reactivation of the developmental program leading to head regeneration [21].

CREB phosphorylation is required for head-regeneration in hydra (Hv). (A) CREB hyperphosphorylation in endodermal cells of the head-regenerating tips (top region, arrows), detected with the anti-phosphoCREB antibody. (B) U0126-treated hydra (20 μM) do not regenerate their head, here shown five days after bisection. The prdl-a-expressing cells (purple points, arrows) indicate the apical pole of the animal, the basal disk appears unstained (arrowheads). Bars: 400 μm. Inlet in A is 4× magnified.
The CREB transcription factor and the RSK kinase are indeed co-expressed in all three hydra-cell lineages including dividing interstitial stem cells, proliferating nematoblasts, proliferating spermatogonia and spermatocytes, differentiating and mature neurons as well as ectodermal and endodermal myoepithelial cells [22]. In addition, CREB gene expression is specifically up-regulated during early regeneration and early budding. Thus, in hydra, the CREB pathway appears already involved in multiple tasks, such as reactivation of developmental programs in an adult context, self-renewal of stem cells, proliferation of progenitors and neurogenesis. More recently, the CREB-Binding protein (CBP) gene was identified, shown to be ubiquitously expressed (SC, unpublished), and is currently tested together with CREB and RSK in RNAi experiments.
2.3 The early–late phase: The Gsx homolog (cnox-2) supports the de novo neurogenesis that precedes head patterning
According to several independent datasets, neurons are supposed to play a minor role in de novo head patterning; for instance, nerve-free hydra mutants can regenerate their heads [23,24]. We recently readdressed this question in wt hydra by testing the function of the ParaHox Gsx-homolog gene, cnox-2. Cnox-2 expression is restricted to fast-cycling interstitial cells that give rise exclusively to sensory mechano-receptor cells (nematocytes) in the body column and apical multipolar neurons. Therefore, cnox-2 is a marker for a subset of interstitial cells that corresponds to bipotent neuronal progenitors [12]. Upon partial cnox-2 silencing, the apical nervous system (ANS), which can be visualized with the anti-β-tubulin antibody, appears reduced and disorganized; when silencing is complete, apical neurons are no longer detected and the body size is drastically reduced (Fig. 4).
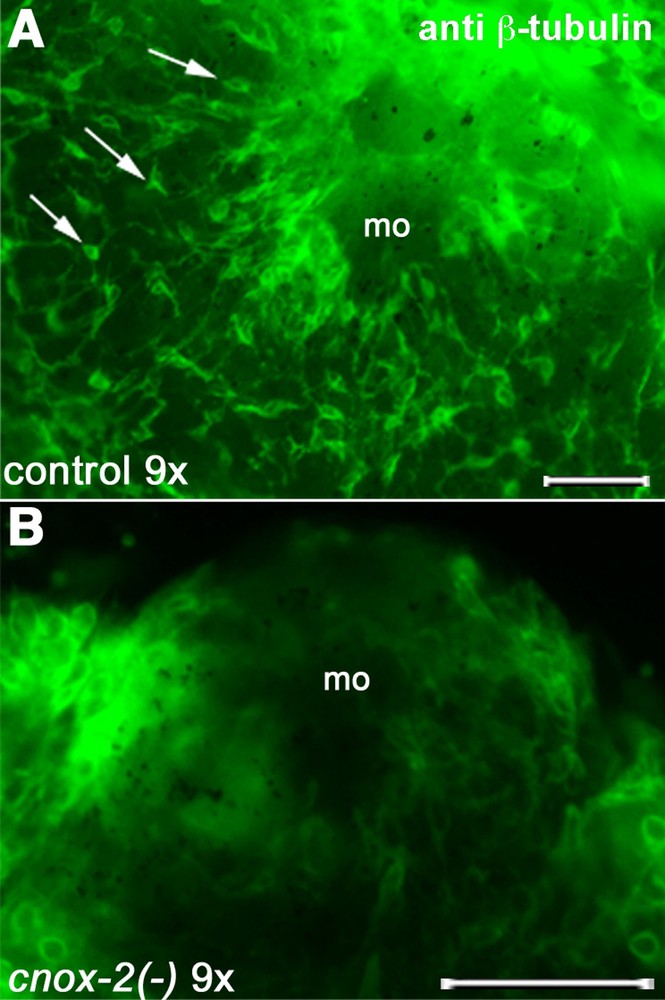
Disappearance of the apical nervous system after nine exposures to cnox-2 dsRNAs. Arrows indicate neurons on control hydra; mo: mouth opening. Bars: 50 μm.
During regeneration, a massive de novo neurogenesis was observed in the head-regenerating tip, starting 24 h post-amputation. The cnox-2 expressing cells, which start to appear in the presumptive head region at the same time, correspond to dividing neuronal progenitors and differentiating neurons. As anticipated, cnox-2 RNAi knockdown alters de novo apical neurogenesis and delays significantly head formation. Similarly, in the sf-1 nerve-free temperature sensitive mutants [25], cnox-2 expression is abolished at restrictive temperature and head regeneration is far less efficient, as 50% of the animals remain unable to regenerate their heads after six days. These results indicate that de novo head patterning in wt hydra polyps depends on cnox-2 promoted neurogenesis [12]. Alternatively, when neurogenesis is missing, a slower and less efficient head developmental program is possibly activated [2].
3 Regeneration in evolution: themes and variations
3.1 Morphallactic versus epimorphic, with or without a blastema?
What is the role of stem cells in the cellular remodelling underlying regeneration? The classical views of regeneration imply a clear distinction between morphallactic regeneration, occurring in the absence of any cell proliferation, and epimorphic regeneration, relying on the formation of a proliferating blastema [26,27]. The first type would concern mostly hydra, and partially planarians [28], whereas the second would correspond to vertebrate regeneration. The recent results obtained in our laboratory suggest that the endodermal myoepithelial cells of the tip undergo a transient phenotypic transition, i.e. they loose their epithelial polarity, while the interstitial cells located immediately underneath re-enter the cell cycle (SC, unpublished). The combination of these two cellular events is highly reminiscent of the blastema formation in urodele regenerating limbs [29] or zebrafish regenerating fins [30]. If confirmed, this would suggest that hydra regeneration in wt conditions might be closer to epimorphic regeneration than anticipated, sharing some regenerative mechanisms at work in amputated urodeles limb or zebrafish fin.
3.2 Nerve dependence of the head regeneration process?
One key aspect in the control of the blastema growth is the presence of neurotrophic factors that play an essential function in the urodele limb [31]. Nerve-free hydra obtained either chemically or genetically provide a useful context to test the nerve dependence. Indeed, those nerve-free hydra can regenerate their head, although with a much weaker efficacy and at a slower pace. At the early phase of head regeneration, the proliferating zone in wt hydra is located in an area that is neuronal rich (SC, unpublished). Therefore, the putative neurotrophic function of these neurons could be tested by comparing the cellular remodelling and the expression profiling in wild-type and nerve-free contexts. At the early–late stage, a de novo neurogenesis was observed at the regenerating tip, which is the site of an intense concomitant cell proliferation. Again, a functional link between these two cellular events is not established. Nevertheless, in nerve-free hydra, an intense proliferation of the myoepithelial cells was also observed at the early–late stage [32]. Are the signals that trigger this cell proliferation in both contexts identical? Are the nerve-free hydra turning on an alternative pathway? Is there any analogy between the nerve-free hydra and the urodele aneurogenic limb [33], i.e. a limb that develops in the absence of any neuronal support and later on does not require any neurotrophic factors for its regeneration? Thanks to the functional tools now available in the hydra model system, these questions should be reconsidered in qualitative and quantitative terms at both cellular and genetic levels.
3.3 Regeneration and developmental plasticity, a long-term memory?
In the coming years, functional studies will tell us how similar are the molecular and cellular processes that drive regeneration in hydra, planarians, annelids, urodeles, and zebrafish. This aspect of regeneration is currently a dark box. To achieve a full understanding of the regenerative potential of adult tissues, their respective regenerative programs have to be elucidated. Nevertheless, more importantly, it will be necessary to understand the permissive context(s) that keeps accessible such developmental programs that de novo give rise to appropriate form and function. Then, a comparative view at the level of gene regulation will be a first step towards the identification of the key regulators of regeneration. The hydra model system provides a unique entry point to identify what does animal regeneration indeed mean, i.e. the capacity to use a minimal number of cellular processes that, given the cell types available, will reactivate a developmental program, which ultimately leads to the reestablishment of the missing part, identical to the amputated one. Surprisingly, the CREB pathway, which was identified as a key signalling pathway for consolidating long-term memory in aplysia, drosophila, and mammals [34], appears as an essential player in the reactivation of the developmental program in hydra. Although we anticipate from our preliminary analyses that this reactivation process in hydra is likely not unique, but rather multiple, it is tempting to speculate that regeneration might reflect the potential for long-term memory mechanisms of developmental processes.
Acknowledgements
This work was supported by the Swiss National Foundation, the Canton of Geneva, the ‘Fonds Georges-et-Antoine-Claraz’ and the Academic Society of Geneva.


