1 Introduction
Ferritin is the most common and ancient molecule of iron homeostasis, essentially ubiquitous and expressed in most eubacteria, archea, plants and animals. The ubiquitous ness and conservedness of ferritin are attributed to its essential role in the control of iron homeostasis and oxygen chemistry [1,2]. Iron (Fe) is an essential nutrient on account of its indispensable function in many fundamental cellular processes. However, in the presence of oxygen, iron can catalyze the production of reactive oxygen species (ROS), which is greatly dangerous for the cell [3].
The typical ferritin is a spherical protein composed of 24 subunits, which fold in a 4-helical bundle. The major property of the protein is the large cavity, designed to accommodate up to 4000 Fe atoms. This very high iron-binding capacity concentrates iron in a compact and safe form, which can be made readily available when needed; therefore, ferritin acts as an iron storage molecule to avoid loss of the precious metal [4].
Most eukaryotes have two major ferritin genes that encode subunits with different properties, generally named H (heavy) and L (light) that co-assemble to form heteropolymers. In mammals, the two subunit-types have little propensity to form homopolymers, and assemble in different proportions thus, originating a large number of isoferritins, with a tissue-specific distribution [1,5]. Thus, L-rich copolymers predominate in spleen and liver, whereas H-rich ferritins are found in other tissues, such as heart and kidney. In heteropolymeric ferritins, the H and L subunits have distinct and complementary functions. The H subunit contains the four-helix bundle a dinuclear ferroxidase center, which promotes the oxidation of Fe(II) in the presence of molecular oxygen [6]. The ferroxidase center sequesters Fe(II) from Fenton-like reactions in which the spontaneous oxidation to Fe(III) donates single electrons to transform innocuous reactive oxygen species, like H2O2, into highly toxic radicals. The iron ligands are highly conserved and are provided by residues E27, E61, E62, H65, E107 and Q141 [7]. The L-chain lacks the catalytic center, but contains specific carboxylic groups on the cavity surface (E57, E60, and E64 using the H-chain numbering), that enable an efficient process of nucleation and mineralization of Fe(III), facilitating iron accumulation [8,9]. As a result, ferritin, through its ability to uptake, oxidize, and sequester iron, reduces free iron level and relieves oxidative stress [10]. The H-rich ferritins have a high ferroxidase activity, therefore, they oxidize and sequester actively the iron and have a more pronounced anti-oxidant activity, while the L-rich ferritins form molecules physically more stable which may contain a larger amount of iron in the cavity and have a more pronounced iron storage function [4,11].
Ferritins from lower vertebrates received relatively little attention. Three distinct ferritin cDNAs, named H (heavy), M (middle) and L (light) on the basis of their mobilities in denaturing gels, have been described in bullfrog tadpole erythrocytes [12]. With respect to the sequence elements of functional importance, the L-chain contains the negatively charged residues responsible for iron nucleation and mineralization in the mammalian protein, the H-chain contains the ligands of the ferroxidase center, and the M subunits possess both the ferroxidase center of H subunits and the iron nucleation site of L subunits [4].
In fish ferritins, evidence for two or three subunits was obtained. The H and M subunits contain both the ligands typical of the mammalian H-chain ferroxidase center and the L-chain carboxylate residues in positions 60 and 64. The canonical L-chain glutamate residue in position 57 is present only in the M-chains and is substituted by an asparagine in the H-chains [13–17]. Evidence for the existence of ferritin L-chains in fish is scanty, since the only data available refer to the amino acid composition of the protein isolated from rainbow trout [18].
Recently, many nucleotidic sequences of H-chain ferritins were characterized from Antarctic teleosts [19–21]; on the contrary, sequences of the M-chain ferritins from Antarctic teleosts were only from the direct amino acid sequencing of the native proteins [15,22]. No L subunits were found in Antarctic fish. The structural and functional properties of the protein extracted from Antarctic fish tissues demonstrated that the presence of a ferritin homopolymer is a typical feature of Antarctic ferritin molecules, since it has been observed that a ferritin homopolymer, made of only M-chains, is present in the spleen of the notothenioids Trematomus newnesi and Trematomus bernacchii [15,22] and a homopolymer made only of H-chains in the spleen of the bathydraconid Gymonodraco acuticeps [23]. However, in both notothenioids, liver ferritin is a heteropolymer composed of M and H subunits [15].
The detection by northern blot analysis of a small but appreciable amount of H-chain mRNA in T. bernacchii spleen suggests the existence in Trematomus spleen of a minor ferritin isoform containing also the H-chain subunit [19]. Indeed, it has been demonstrated that the two subunit-types together improve the flexibility and functionality of the molecule [1].
The main objectives of this work were to increase the scarce molecular data on M-chain ferritins in Antarctic fish, and to provide new insights about the evolution of ferritin subunits in fish. To this end, a molecular characterization study has been made of the ferritin M-chains in the Channychtydae species Chionodraco rastrospinosus. Results report the identification of five different ferritin M-chain cDNA sequences. Gene expression analysis demonstrates that transcripts encoding both H and M subunits are present in all the tissues examined, whereas no L-chains have been found. Finally, a phylogenetic analysis was inferred to explore the evolutionary mechanisms of ferritin in fish.
2 Materials and methods
2.1 Animals
Adult Chionodraco rastrospinosus specimens were collected in the proximity of the Antarctica Peninsula and kept in aquaria supplied with aerated seawater at approximately −1.5 °C. Animals were anaesthetized in 0.50 g/L MS-222 (tricaine methane sulphonate) and then killed by severing their spinal cord behind the head plates. Tissues were quickly removed, frozen in liquid nitrogen, and stored at −70 °C until use.
2.2 Total RNA extraction
Total RNA was isolated from tissue samples using the TRI Reagent RNA Isolation Reagent (Sigma–Aldrich) according to the manufacturer's instructions, and resuspended in diethyl-pyrocarbonate-treated water. After removal of contaminating DNA using the TURBO DNA-free kit (Ambion), the concentration and integrity of the RNA samples were determined by absorbance at 260 nm and by formaldehyde–agarose gel electrophoresis.
2.3 First-strand cDNA synthesis
Total RNA (5 μg) isolated from fish tissues was reverse-transcribed using an anchored oligo-dT primer (PK71) and the Superscript-II reverse transcriptase (Invitrogen), as previously described [24].
2.4 PCR and 5′-RACE amplification of cDNA
All the PCR and 5′-RACE (Rapid amplification of 5′ cDNA ends) experiments were performed using the Fast Start Taq DNA polymerase (Roche Diagnostics) and a peqSTAR thermal cycler (PEQLAB Biotechnologie). PCR reactions were carried out on 2 μl of first-strand cDNA, with an initial denaturation step at 94 °C for 4 min, 30 cycles made of a step at 94 °C for 30 s, a step at 50 °C for 30 s, and a step at 72 °C for 1 min, followed by a final elongation step at 72 °C for 10 min. The 5′-ends of the cDNAs were obtained by the RACE protocol of the Marathon cDNA Amplification kit (BD Biosciences) as previously described [25]. PCR conditions were set according to the Marathon kit protocol.
Specific primers used for the PCR and 5′-RACE are listed in Table 1. The primers were designed on the nucleotide sequences of piscine ferritin M subunit available at the EMBL Nucleotide Database. All the primers were synthesized by Primm Laboratories (Italy). The AP1 primer, used for 5′-RACE, was supplied with the Marathon cDNA amplification kit.
Specific primers used for PCR and 5′-RACE analyses.
| Name | Nucleotide sequence (5′-3′) | Length | Direction |
| FerM-Frw1 | TGAATTCGCACTTAAATCCATTC | 23-mer | Sense |
| FerM-Frw2 | TACCGTTTTGTTAAGAAAAACAG | 23-mer | Sense |
| FerM-Frw3 | TCACTTCTTCAAGGAGAACAGC | 22-mer | Sense |
| FerH-Rev2 | ACGCTCTTCTCCAGCTGCAGGG | 22-mer | Antisense |
| FerH-M | GACTGCGAGGCGGCCGT | 17-mer | Sense |
2.5 Cloning and sequencing of PCR products
The purified PCR products were directly cloned into the pSC vector using the StrataClone PCR cloning kit according to the manufacturer's instructions (Agilent Technologies). Several randomly selected colonies were inoculated into LB broth containing 100 μg/ml ampicillin and incubated overnight at 37 °C with shaking. Plasmids containing inserted sequences were purified using Fast Plasmid columns (Eppendorf) and sequenced using automated methods on an ABI PRISM Genetic Analyzer (PE Biosystems). Sequences and annotations of C. rastrospinosus ferritin M-chain cDNAs appear in the DDBJ/EMBL/GenBank database with accession numbers shown in Table 2.
Ferritin M-chain subunits expressed in the Chionodraco rastrospinosus liver.
| Acronym | cDNA length (bp) | 5′-UTR length | 3′-UTR length | CDS length | Number of aa residues | Accession number |
| C.rasM1 | 811 | 133 | 150 | 528 | 176 | HF559476 |
| C.rasM2 | 497 | 177 | n.d. | 320 | 106 | HF559477 |
| C.rasM3 | 506 | 186 | n.d. | 320 | 106 | HF559478 |
| C.rasM4 | 503 | 183 | n.d. | 320 | 106 | HF559479 |
| C.rasM5 | 314 | n.d. | n.d. | 314 | 104 | HF559480 |
2.6 Multiple alignments and sequences analyses
Sequences of fish ferritin subunits were retrieved from the NCBI database. The accession numbers of the selected sequences are reported in Table 3. The multiple alignment of amino acid sequences (Fig. S1, Supplementary data) was obtained using the program ClustalW version 2.1 [26] and was refined manually by the program Se-Al v. 2.0a11.
Source, gene name, accession number and acronym of the 34 amino acidic sequences used for the phylogenetic analysis of fish ferritins.
| Organism | Gene name | Accession number | Acronym |
| Danio rerio | Ferritin H-chain | NP_571660 | DrerH1 |
| Ferritin H-chain | NP_001004562 | DrerH2 | |
| Ferritin H-chain | NP_001002378 | DrerH3 | |
| Ferritin H-chain | XP_001921731 | DrerM1 | |
| Ferritin M-chain | XP_687175 | DrerM2 | |
| Ferritin M-chain | NP_001124139 | DrerM3 | |
| Ferritin M-chain | NP_001018367 | DrerM4 | |
| Ferritin M-chain | NP_001103324 | DrerM5 | |
| Ferritin M-chain | NP_001103175 | DrerM6 | |
| Ferritin M-chain | NP_001107131 | DrerM7 | |
| Ferritin M-chain | AAI71692 | DrerM8 | |
| Ferritin M-chain | AAI64954 | DrerM9 | |
| Ferritin M-chain | AAI61623 | DrerM10 | |
| Salmo salar | Ferritin H-chain | NP_001117129 | SsalH1 |
| Ferritin H-chain | NP_001133194 | SsalH2 | |
| Ferritin H-chain | NP_001139960 | SsalH3 | |
| Ferritin M-chain | NP_001117130 | SsalM1 | |
| Ferritin M-chain | NP_001134896 | SsalM2 | |
| Oreochromis niloticus | Ferritin H-chain | XP_003445743 | OnilH1 |
| Ferritin H-chain | XP_003440525 | OnilH2 | |
| Ferritin H-chain | XP_003438521 | OnilH3 | |
| Ferritin M-chain | XP_003446457 | OnilM1 | |
| Ferritin M-chain | XP_003446470 | OnilM2 | |
| Trematomus bernacchii | Ferritin H-chain | FM210465 | TberH1 |
| Ferritin M-chain | P85836 | TberM1 | |
| Ferritin M-chain | P85839 | TberM2 | |
| Chionodraco rastrospinosus | Ferritin H-chain | AM419433 | CrasH1 |
| Ferritin M-chain | HF559476 | CrasM1 | |
| Ictalurus punctatus | Ferritin H-chain | NP_001187267 | IpunH1 |
| Ferritin H-chain | NP_001232867 | IpunH2 | |
| Ferritin M-chain | NP_001187268 | IpunM1 | |
| Ferritin M-chain | ADO29006 | IpunM2 | |
| Epinephelus bruneus | Ferritin H-chain | AEI87382 | EbruH1 |
| Ferritin M-chain | AEI87383 | EbruM1 |
2.7 Phylogenetic analyses
Phylogenetic trees were inferred by Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian models. MP analysis was performed with SeaView version 4 software [27]; ML analysis was performed with MEGA software v. 5.0 [28]. The WAG + G substitution model was chosen on the basis of the result from ProtTest [29] and MEGA analyses. Bootstrap branch support was estimated using 1000 data sets. Bayesian analysis was carried out using the software MRBAYES v. 3.1 [30] (http://mrbayes.net). Five millions generations were run, and trees were sampled every 100 generations. The resulting trees were summarised in a consensus tree. Tree visualization and final edition was performed in FigTree v. 1.3.1.
3 Results
3.1 Isolation of cDNA clones for icefish ferritin M-chain and sequence analyses
Assembly of the RT-PCR products, obtained with primers designed on the M-chain of zebrafish ferritin, returned for the icefish C. rastrospinosus a complete mRNA sequence, plus four partial mRNA coding sequences, all sharing a clear homology with the M-chain of other fish species. The complete mRNA sequence (C.rasM1) was made of 811 bp with an open reading frame of 528 bp encoding for a protein of 176 amino acidic residues, and 5′- and 3′-UTR of 133 and 150 bp, respectively. The lengths of the partial mRNA fragments are reported in Table 2. In Figs. 1 and 2, the alignments of nucleotidic and amino acidic sequences, respectively, are reported. Although the isoforms share more than 90% identity, one or more differences have been found in the amino acid sequences. Unique among the sequences, the isoform C.rasM4 lacks the canonical glutamate residue in position 54 (57 in the mammalian L-chain), substituted by the non-conservative alanine residue (Fig. 2).
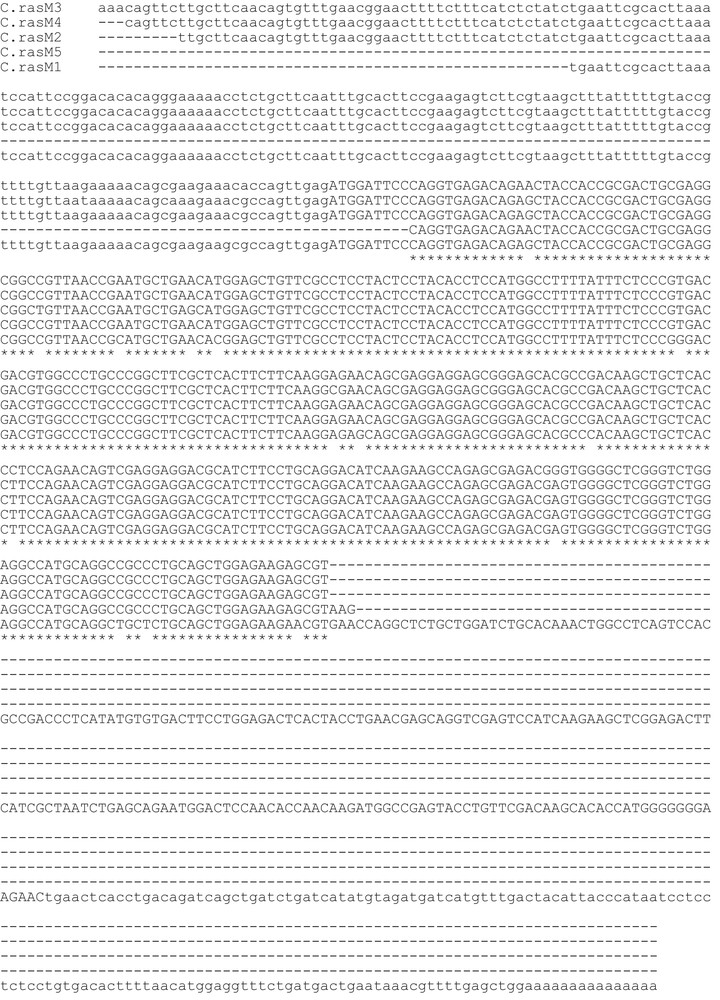
Aligned nucleotide sequences of cDNAs encoding Chionodraco rastrospinosus ferritin M isoforms. The open reading frames are indicated by capital letters, 5′- and 3′-untranslated regions by low-case letters. Identities are represented by asterisks, deletions by hyphens.
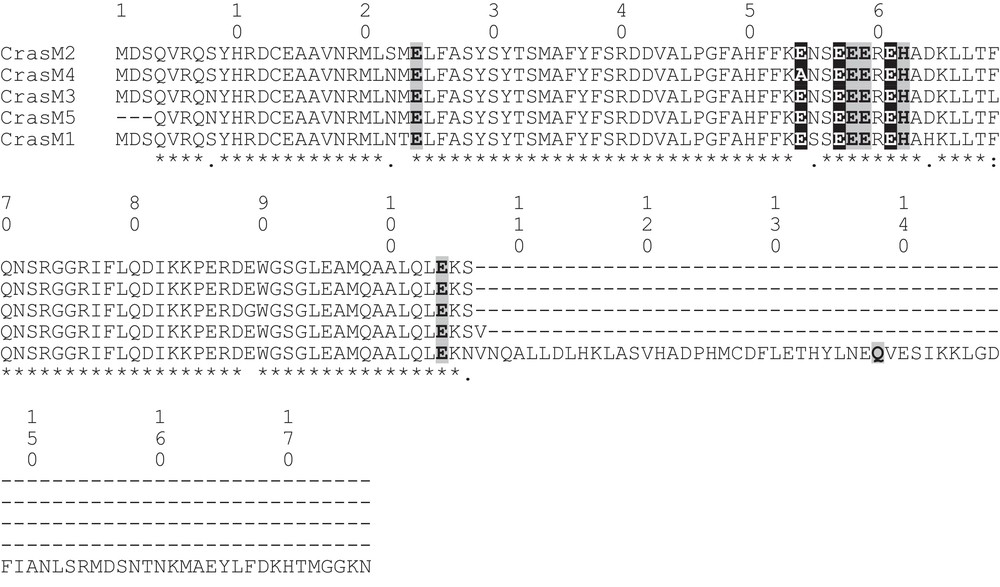
Alignment of the amino acid sequences of ferritin M isoforms from the icefish C. rastrospinosus. The six iron ligands of the dinuclear ferroxidase center and the three carboxylic ligands of the iron nucleation center are shadowed in light gray and dark gray, respectively.
The amino acid sequence of C. rastrospinosus ferritin H-chain was previously deduced from nucleotide sequencing of the corresponding cDNA [19]. Fig. 3 shows a comparison of the amino acid sequences of the C. rastrospinosus H- and M-chain ferritins. The sequences identity was calculated to be 67.8%, similarity was 83.6%. As for other fishes, the M-chain contains both the ferroxidase center and the iron nucleation site; the H-chain shows the ferroxidase center and two of the three residues forming the iron nucleation site. Only a gap is present between the sequences, lacking the 177th residue in the M-chain.

Alignment of the complete amino acid sequences of ferritin H and M1 chains from the icefish C. rastrospinosus. The six iron ligands of the dinuclear ferroxidase center and the three carboxylic ligands of the iron nucleation center are shadowed in light gray and dark gray, respectively. Asterisks indicate identity between residues; colons and dots indicate conservative and semi-conservative amino acid residues, respectively.
3.2 Expression of H- and M-chain transcripts in C. rastrospinosus tissues
In order to establish the presence of both the H and M ferritin subunits in different tissues, PCR reactions were carried out using a ferritin specific primer, the 17-mer FerH-M (Table 1), matching exactly with all the H and M isoforms, and as antisense primer a non-specific oligonucleotide corresponding to the oligo(dT) primer used in the first-strand synthesis, but lacking the oligo(dT) tail [24]. Electrophoretic analysis of the PCR products retrieved the presence of two transcripts, one of about 700 bp, corresponding to a ferritin H-chain cDNA fragment, and another of about 600 bp, corresponding to a ferritin M-chain cDNA fragment. The nature of the two transcripts was confirmed by automatic sequencing. The two transcripts were found in all the tissue examined (Fig. 4). The analysis was repeated on tissues from three different C. rastrospinosus specimens.

Detection of H and M ferritin transcripts in C. rastrospinosus tissues by conventional RT-PCR. The RT-PCR analysis was carried out as described under methods and the resulting products analysed on a 1.2% agarose gel stained with ethidium bromide. The 700 bp band corresponds to the H-chain cDNA fragment, the 600 bp to the M-chain cDNA fragment. Lane 1:100 bp DNA ladder; lanes 2–6: RT-PCR products from C. rastrospinosus blood (2), kidney (3), spleen (4), head kidney (5) and liver (6). The RT-PCR analyses were performed on three sets of tissues from three different C. rastrospinosus specimens.
3.3 Phylogeny of H and M ferritin subunits in fishes
Thirty-four amino acid sequences of H and M ferritin isoforms from different fishes were aligned and used to derive new insights on the evolutionary history of fish ferritin subunits. The accession numbers of the final set of sequences are listed in Table 3. The alignment is reported in Fig. S1, Supplementary data of the online version of this paper. The phylogenetic analysis of the dataset was carried out according to three different methods, namely ML, MP and Mr Bayes, to avoid possible inconsistency due to the use of a single method. The maximum likelihood tree is shown in Fig. 5. The support for the different branches was determined by bootstrap analysis.
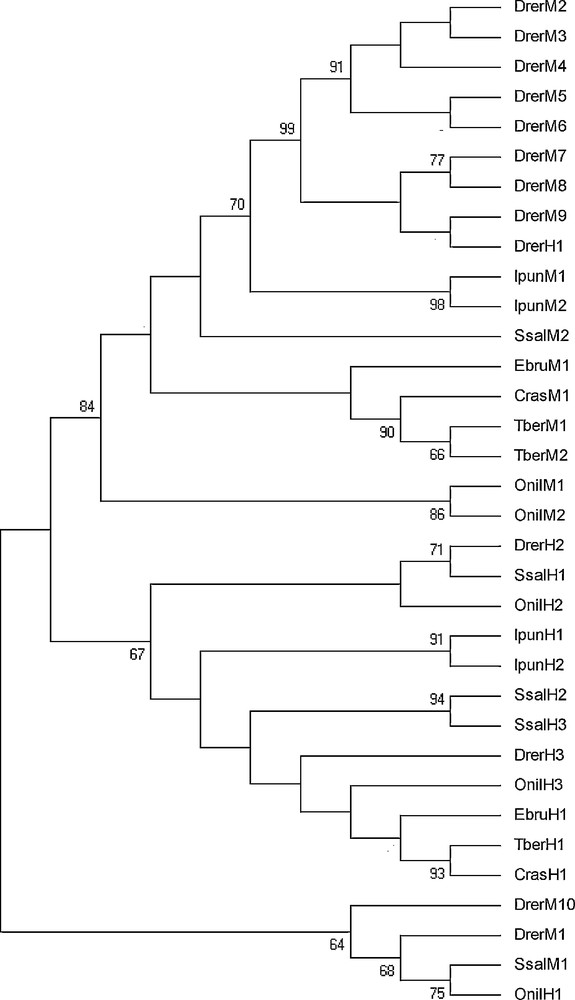
Phylogenetic tree of the amino acid sequences of the piscine ferritin H and M isoforms inferred by the Maximum Likelihood (ML) method using the WAG+G substitution model implemented in the software MEGA v. 5.0. The same topology was obtained by Maximum Parsimony and MRBAYES (Materials and methods). At the nodes are given values of ML non-parametric bootstrap (cut-off 65%). For full species name, refer to Table 3.
Trees identified two major clades, one of ferritin H isoforms and another comprising ferritin M isoforms. This topology was strongly supported by all the methods used, providing evidence that diversification of piscine ferritin chains occurred before the separation among piscine orders. Nevertheless, the ferritin M-chains clade also comprises a D. rerio ferritin H-chain isoform. Basal to these two clades, from all the analyses emerges another quite heterogeneous clade, formed by two D. rerio ferritin M isoforms, a S. salar ferritin M and an O. niloticus ferritin H isoform.
4 Discussion
In the process of cold adaptation, the evolutionary trend of Antarctic fish has led to unique specializations in many biological features in comparison with temperate and tropical species [21,31].
The most striking adaptive character of Antarctic Notothenioidei, the dominant component of the Southern Ocean fauna, is the reduction of erythrocytes number and haemoglobin (Hb) concentration [32]. In the Channichthyidae family, the blood pigment is absent [33]. Reductions of the haematocrit appears selectively advantageous because it diminishes the energetic cost associated with circulation of highly viscous, corpuscular blood fluid [34]. On the other hand, because oxygen solubility in seawater is inversely proportional to temperature, the cold Antarctic Ocean is an exceptionally oxygen-rich aquatic habitat. So, icefish maintain normal metabolic function by delivering oxygen physically dissolved in blood, which has an oxygen carrying capacity of less than 10% of that seen in red-blooded notothenioid fishes [35].
This unique peculiarity among vertebrates has made this fish group a suitable system to studying the relationship between Hb and the other iron-binding proteins and to understand the adaptive changes occurred in Antarctic fish metabolism to avoid the deleterious effects of iron overload in the absence of haemoglobin.
Recent evidence demonstrated that the loss of Hb in icefishes is paralleled by the loss of iron in blood cells and plasma fractions [19]. Nevertheless, the intake of iron in these fish occurs by food, consisting generally in red-blooded, iron-rich fishes. So, in icefish, a mechanism must be for controlling the homeostasis and keeping the iron in the tissues in a non-toxic form. Indeed, recent investigations on the expression of key genes involved in metabolism, transport and storage of iron have shown that these genes, in contrast to what happens to the Hb gene [36], are fully functional, although their expression is regulated differently in various tissues [19–21,37]. In particular, transferrin and ceruloplasmin genes show no significant differences in the expression between red-blooded and hemoglobinless Antarctic fish, whereas genes encoding the ferritin and the divalent metal transporter are expressed to a lesser extent in the hematopoietic organs of icefishes. However, studies on ferritin gene expression in Antarctic fishes regarded only the H-chain [19,20], being the ferritin M-chain investigated only at the protein level [15,22]. Data here reported demonstrate that in the icefish C. rastrospinosus the ferritin M-chain transcript is constitutively present in all tissues examined, as other teleosts, the gene is present in multiple isoforms. On the contrary, no multiple isoforms of ferritin H-chain are found in the Antarctic fish so far analyzed [20]. The presence of a detectable amount of ferritin transcripts in blood, accompanied by a concomitant absence of iron in the same tissue, suggest that in C. rastrospinosus the ferritin mRNAs may not be translated; alternatively, the protein may be present as apoferritin. The presence of both H and M transcripts in the icefish tissues demonstrates that the presence of ferritin homopolymers in Antarctic fish tissues is not due to the absence of one of the two transcripts, but rather could be due to the functional characteristics of the two different chains. In particular, the increased functionality and flexibility observed by ferritin heteropolymers may not play a major role for animals in which iron accumulation in certain tissues is not a priority. In addition, it must be considered that the Antarctic fish, as well as other teleosts, possess a ferritin M-chain endowed of both the ferroxidase center typical of H subunits and the iron nucleation site typical of the mammalian L subunits. The maintenance of both functions leads to the assumption that the M-chain could represent the most ancestral form of ferritin, from which had arisen the H-chain by gene duplication. Indeed, in phylogenetic trees inferred in this study, ferritins are separated in two clades, one grouping the H-chains and another that includes the M-chains, with high bootstrap support. However, a cluster consisting of two M-chain isoforms from D. rerio, one M-chain isoform from S. salar and one H-chain isoform from O. niloticus merges basal to the two major clades. This would suggest that some piscine species have retained ancestral forms of ferritin, which may have given rise to the more specialized H- and M-chains. The presence in the latter clade as well as in the clade clustering the M-chain of an H-chain, might be due to the very scarce differences showed by the two ferritin subunits in fish.
Despite significant advances in our understanding of ferritin subunits in fishes, it is evident that further work is needed. As regarding phylogenetic data, future efforts should be centered on obtaining data for other piscine orders, including DNA sequences, not always available. In addition, another important goal should be to infer a phylogenetic tree of ferritin molecules involving more taxonomic groups of vertebrates.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.


