1 Introduction
Agaricus bisporus (Lange) Imbach, the button mushroom, is a leaf-litter rot fungus. Of the major cultivated mushrooms, it has until recently been the least well known from nature. Since 1990, numerous isolates have been collected throughout the world at different sites on different kinds of decomposing lignocellulosic materials, and three varieties having different life cycles have been identified [1,2]. For many years, the button mushroom has been grown in caves at 15–16 °C. Since the 1980s, climatic chambers replace caves, and cultivation conditions are standardized, with a 16–18 °C temperature recommended during the cropping period, based on the requirement of the cultivated strains. Thermo-tolerance is an interesting trait to be studied, both for the cultivation of the mushroom out of temperate countries, and for the understanding of the distribution of wild populations under various climates. Usually, a temperature over 32 °C stops the growth of the vegetative mycelium. In a recent work, 91 isolates from various geographic areas were screened for their ability to fruit at higher temperatures (FHT+ strains fruiting at 25 °C) than the commercial cultivars. The mushroom variety discriminated for this trait. Agaricus bisporus var. eurotetrasporus was unable to develop any sporophore whilst A. bisporus var. burnettii was well adapted to fruit both at 17 and 25 °C, suggesting this phenotypic plasticity is a fixed trait in this variety. In the more common A. bisporus var. bisporus, the ability to fruit at 25 °C was observed in some strains, but it correlated neither with climate/microclimate nor with habitat of the strains, and yields were always lower than at 17 °C [3]. However, we do not know the temperature at which the ancestor of the species fruited, neither the order in which the three varieties were separated.
To date, information on thermo-tolerance mechanisms of A. bisporus is scarce. Another cultivated mushroom, Flammulina velutipes (Curtis) Singer is generally regarded as a fungus that requires a low temperature (< 15 °C) for the induction or production of fruiting bodies, as compared with vegetative growth, which has an optimum temperature of 22 to 26 °C [4,5]. Fultz [6] analyzed progenies of five crosses between strains with high and low fruiting temperature indicating that a minimum of two genes appeared to control the requirement for fruiting at > 15 °C. Mutants able to fruit at 20 °C and, for most of them, to grow at 33 °C were recently obtained by ultraviolet irradiation mutagenesis and combined mutagenesis [7]. In filamentous Ascomycota fungi, over-expression of a heat shock proteins (HSP70) enhances thermo-tolerance of Trichoderma harzianum [8]. Heat shock factor proteins (HSFs) are transcriptional regulators of genes that encode different types of stress proteins as HSPs. In Coniothyrium minitans, HSF1 over-expression enhances tolerance to heat stress [9].
For A. bisporus, Chen [10] obtained few thermo-tolerance-related gene fragments with DD-RT-PCR to analyze gene expression in the vegetative mycelium of one A. bisporus strain (02) under normal and higher-temperature cultivation. They identified 3 genes of thermo-tolerance: 028-1, 023-11A and 023-11B [10–14]. The full-length cDNA sequence of one gene (028-1 GenBank accession number DQ235473) had been obtained [13,15]. It has a size of 1.37 kb, and no obvious homological sequence was found in GenBank. The Chinese group constructed a binary expression vector of 028-1 of A. bisporus and transferred the gene into the non-thermo-tolerant strain 8213 of A. bisporus by Agrobacterium-mediated transformation. Eight out of ten of transgenic strains were able to grow at 34 °C as well as the thermo-tolerant strain from which 028-1 had been identified, whereas the wild strain did not [14]. However both in the thermo-tolerant parent and the transformants, the mycelial growth rate was strongly affected, with mycelial diameters after 22 days at 34 °C being 15 to 25% that at 28 °C. Despite the potential interest of this gene and its regulation, no other article than those published by Chen et al. was available. Therefore we proposed that an improvement of its annotation, a study of its polymorphism and level of expression in different strains should allow characterizing 028-1 and determine accurately the role of this gene in thermo-tolerance of A. bisporus.
The aim of the present work was then to progress in the knowledge of 028-1 gene and its involvement in the thermo-tolerance of some A. bisporus strains. The level of expression of 028-1 in the vegetative mycelium during a heat treatment was analyzed by Q-PCR on several strains. We identified and annotated 028-1 in the genome of the homokaryon of the hybrid HU1 (H97) that had been used for the whole genome sequencing of A. bisporus var. bisporus (http://genome.jgi-psf.org/Agabi_varbisH97_2/Agabi_varbisH97_2.home.html), but this hybrid is not thermo-tolerant. We established that 028-1 is a homolog of the YAP1 gene involved in oxidative stress response in yeast.
Yap1p is an AP-1-like transcription factor. AP-1 is an activator protein that links to the promoter of the human metallothionein gene and the simian virus 40 (SV40) [16]. Yap1p is one of the 15 basic-leucine Zipper (bZIP) transcription factors implicated in various forms of stress response in yeasts. These are clustered in 7 families, 4 conserved in metazoans (Atf2, Atf6, CREB1 and Jun) and 3 only found in yeast (Yap, Met4 and Met28). The YAP (yeast activator protein) family includes eight members (Yap1-Yap8) [17–19]. Three of these members (Yap1, Yap2 and Yap8) contain Yap1 redox domain (IPR023167). A. bisporus 028-1 protein also contains this domain but differs from Yap1P in his sensing capability. That is why we propose to name aap1 the gene 028-1.
Data on 028-1 polymorphism and level of expressions in primordia and sporophores obtained at two temperatures in different strains are presented and discussed on the light of its putative role in thermo-tolerance of A. bisporus.
2 Materials and methods
2.1 Strains of Agaricus bisporus
The strains used in the different parts of the work are presented in Table 1. They all are maintained in Collection Germplasm of Agaricus at Bordeaux (CGAB) INRA, France.
Strains of A. bisporus: origin, thermo-tolerance phenotypes, aap1 genotypes and deduced protein phenotypes.
| Strains of A. bisporus | Isolation area | Fructification at high temperature (25 °C) | Survival of mycelium at 33 °C | Genotypes, nucleic acids | Amino acids | |
| Bs0004 | var. bisporus | Canada – British Columbia | 0a | 0 | 4/5 | 2/2 |
| Bs0085A | ” | France – St-Germain-des-Essourts | 0 | 0 | 3/4 | 1/2 |
| Bs0190 | ” | France – West- Lorient | 1 | 0 | 1/4 | 1/2 |
| Bs0285 | ” | France – West- La Rochelle | 1 | 0 | 2/4 | 2/2 |
| Bs0370 | ” | France – West- St Malo | 0 | 0 | 1/4 | 1/2 |
| Bs0374 | ” | England | 1 | 0 | 1/1 | 1/1 |
| Bs0416D | ” | France – Guesnes | 0 | 0 | 1/4 | 1/2 |
| Bs0419B | ” | France – South West- Capbreton | 1 | 0 | 1/5 | 1/2 |
| Bs0420 | ” | France – South West - Vieux-Boucau-les-Bains | 0 | 0 | 1/3 | 1/1 |
| Bs0431C | ” | France – West- La Rochelle | 1 | 0 | 1/11 | 1/2 |
| Bs0470 | ” | Russia – Penza | 1 | 0 | nd | nd |
| Bs0474 | ” | Russia – Penza | 1 | 0 | 1/4 | 1/2 |
| Bs0483 | ” | Canada – Alberta | 1 | 0 | 6/7 | 1/3 |
| Bs0533 | ” | Cultivar – white hybrid, Amycel 2100 | 0 | nd | nd | nd |
| Bs0564 | ” | Greece – Rovies | 0 | 1 | 4/11 | 2/2 |
| Bs0571 | ” | Greece – Larissa | 1 | nd | 15/16 | 1/2 |
| Bs0572 | ” | Greece – Crete Island- Zourva | 0 | 0 | 2/4 | 2/2 |
| Bs0584 | ” | Italy – Sicily | 1 | 0 | 5/12 | 2/2 |
| Bs594 | ” | Greece – Limni | nd | nd | 17/? | 2/? |
| Bs0661 | ” | Greece – Gravia | 1 | 0 | 1/4 | 1/2 |
| Bs0679 | ” | Spain – Sangüesa-Aragon | 0 | 1 | 1/1 | 1/1 |
| Bs0705 | ” | Portugal – Alentejo | 1 | 0 | 13/14 | 2/2 |
| Bs0717 | ” | USA – New Mexico | 0 | 0 | 6/7 | 1/3 |
| Bs0739 | ” | Mexico – Tlaxcala | 1 | 1 | 1/6 | 1/1 |
| Bs0094 | var. burnettii | USA – California- Sonoran Desert | 1 | 0 | 8/8 | 1/1 |
| Bs0738 = Jb137 |
” | USA – California- Sonoran Desert | 1 | 0 | 9/10 | 1/4 |
| Bs0423 | var. eurotetrasporus | France – West- La Rochelle | 0 | 0 | 4 | 2/2 |
| Bs0514 | ” | Greece – Larissa | 0 | 0 | 4 | 2/2 |
| Bs0261 | Hybrid var. bisporus and eurotetrasporus | France – St Malo Dinard | 0 | 0 | 1/4 | 1/2 |
| Bs0026 | var. bisporus | Cultivar – white hybrid, Sylvan HU1 | 0 | 0 | 1/4 | 1/2 |
| Bs0026-7 | Homocaryon var. bisporus | Cultivar – white hybrid, Sylvan HU1 | 4 | 2 | ||
| H97 | Homocaryon var. bisporus | Cultivar – white hybrid, Sylvan HU1 | 1 | 1 | ||
| Jb137-s8 | Homocaryon var. burnettii | USA – California- Sonoran Desert | 9 | 4 |
a 0 = did not produce fruiting bodies at 25 °C in Largeteau et al. [3].
2.2 Thermo-tolerance of the vegetative mycelium
In a first experiment we selected eight wild strains from our previous work [3], based on their ability (FHT+) or inability (FHT−) to produce mature sporophores at 25 °C and to survive an exposure to 33 °C. They were five A. bisporus var. bisporus (Bs0483, Bs0533, Bs0679, Bs0571, and Bs0739), two A. bisporus var. burnettii (Bs0094, Bs0738) and a natural hybrid between var. bisporus and var. eurotetrasporus (Bs0261). They had been collected under sub-tropical, temperate or cold climates in America and Europe. Each strain was cultivated on liquid Cristomalt medium (Barley malt powder, Dif.A1) for 14 days at 25 °C in darkness and then divided into two samples. For each strain three samples were continuously cultivated at 25 °C, and three others were transferred at 32 °C for 12 h to induce expression of the thermo-tolerance-related genes. Mycelium was harvested immediately, washed with iced water and immediately placed in liquid nitrogen to stop the synthesis of mRNA before being stored at −80 °C until RNA extraction and purification.
In a second experiment, 25 wild strains of A. bisporus and a homokaryon (Bs0026-7 = HU 1-7), obtained in our lab by P. Callac [20] from the cultivar hybrid HU1, were studied. Two of them were A. bisporus var. burnettii, two were A. bisporus var. eurotetrasporus (Bs0423, Bs0514), 20 were A. bisporus var. bisporus and one was the natural intervarietal hybrid (Bs0261). Their survival and growth ability were tested here at 18 °C, 25 °C, 30 °C and 33 °C. Inoculum plugs (5 mm diameter) were removed from the edge of 15-day-old cultures and placed in the center of Petri dishes filled with Cristomalt agar medium. Three replicates were observed by strain and scored for survival and mycelial growth rates.
2.3 Production of primordia and fruiting bodies at 25 °C
Nine wild strains from CGAB (INRA, France) previously identified for their ability to produce mature fruiting bodies at 25 °C [3] were cultivated at an experimental scale. Spawn was prepared with cooked rye grains purchased by Euromycel (France) and cultivation was in 0.09 m2 crates filled with 8 Kg of compost purchased from a local mushroom grower (Pons, France) and spawned at 0.8%. Five replications were used. Incubation lasted 13 days at 25 °C. After casing and post incubation during 7 days, half of the crates were moved to a fruiting room regulated at 17 ± 2 °C and 87–90% relative humidity in air; the other half to a fruiting room regulated at 25 ± 2 °C and 92–95% relative humidity. Primordia and fruiting bodies produced at 17 °C or 25 °C were harvested in one crate for each strain and put immediately on ice. The pilei-pellis of fruiting bodies and gills were removed from the fruiting bodies which were placed in liquid nitrogen to stop the synthesis of mRNA and stored at −80 °C until being required for RNA extraction and purification. The sampling units for RNA extractions were pools of extracts obtained from 25 primordia, or from five sporophores. The level of production of sporophores, for each strain, was estimated at each temperature on four crates.
2.4 Expression analysis of 028-1
Total RNA was extracted from samples of mycelia, primordia and sporophores stored at -80 °C. The frozen biomass was crushed with liquid nitrogen. Lyse of cells was obtained with the QIA shredder™ kit from Qiagen. The extraction and the purification of RNA were performed with the RNeasy® Minikit (Qiagen) according to manufacturer's instructions with slight modifications: two stages of digestion of the genomic DNA were added, using the RNase-Free DNase Set (QIAGEN). The first one was during the extraction and the second during the purification. The final concentration and purity of extracted RNA were quantified using the absorption ratio of A260/A280 and A260/A230 measured with NanoDrop® ND-1000, Labtech. Aliquots of total RNA were stored at −20 °C.
RT-PCR was carried out using OneStep RT-PCR Kit (QIAGEN) with 800 ng of total RNA and 0.5 μM of oligo (dT) (Promega) for a final volume of 14 μL for each tube, in a PCR Thermocycler (Master cycler, Eppendorf). The samples were then incubated during 10 min at 65 °C and put in ice.
The following reaction mixture was added for each tube: 2.5 μL DTT, 5 μL 5X first strand buffer, 1 μL M-MLV-RT (Kit M-MLV reverse transcriptase, Invitrogen), 2 μL dNTP, 0.5 μL RNasin ribonucleic inhibitor (Promega), for a 25 μL final volume. The tubes were incubated for one hour at 37 °C and 15 min at 70 °C. cDNA concentration was measured by spectrophotometry at 260 nm by using Nanodrop® ND-1000 (Labtech). For every strain and every condition, aliquots of 20 ng/μL cDNA were stored at −20 °C.
EF1-α (GenBank X97204) was used as housekeeping gene, with the primer pair EF3F (5′-TGGTCGTGGTGAGACTGGTA-3′) and EF2R (5′-GGGTCGTTCTTGGAATCAGA-3′). For 028-1 (GenBank FJ940724.1 thermo-tolerance gene) the primer pair was Th5F (5′-AAGTGGGATGCTACCAAGG-3′) and Th5R (5′-GGATATGGAACTCCACAGCG-3′). Real-time PCR was performed using a LightCycler® 1.5 PCR (Roche Diagnostic) and the Light Cycler® Fast Start DNA MasterPLUS SYBR Green I (Roche) kit, according to the manufacturer's recommendations. Every capillary contained 20 μL of reaction mixture which included 1 μL of primer sense (500 nM), 1 μL of primer antisense (500 nM), 4 μL of Master's Mix, 9 μL of water PCR-grade deionized and 5 μL of cDNA (2 ng/μL). All PCR reactions were carried out in triplicate under the following conditions: a step of enzyme activation at 95 °C during 10 min, followed by 40 cycles of amplification (denaturation at 95 °C during 15 s; annealing at 59 °C during 15 s; extension at 72 °C during 20 s), and finally, a cooling step at 40 °C for 30 s to ensure that the thermal chamber has cooled down upon opening the lid. The experiment was performed three times. A melting curve was run at the end of the 40 cycles to verify for a unique PCR reaction product. Standard curves were built with seven serial dilutions of cDNA pools and each set of primers to check for identical amplification efficiency with material of the strain Bs0470.
In order to determine the gene expression level, we performed a relative quantification analysis using the comparative SYBR Green/ΔCT method. Results were normalized using ΔCT values obtained from housekeeping control gene ef1-α amplification products included in the same runs. All targets were analyzed in triplicate and mean levels of gene expression were calculated. An estimation of relative's levels of transcription of 028-1 gene was performed by Pair Wise Fixed Reallocation Randomization test on CT values using the REST-384® software. Six measures per sample and condition were used for statistical analysis. The ratios (R) are calculated between the target gene (028-1) expression and the reference gene (ef1-α) according to the formula:
- • R is the relative expression ratio of a gene in a sample/a control
- • Exx is PCR amplification efficiency of the gene
- • CTxx is the threshold cycle of the PCR amplification of the gene xx.
2.5 Identification of homologs, sequences alignment and annotation of 028-1 gene
BLASTn was used for identifying 028-1 gene in the genome of A. bisporus var. bisporus (H97) and A. bisporus var. burnettii (JB137-S8) available at http://genome.jgi-psf.org/Agabi_varbisH97_2/Agabi_varbisH97_2.home.html and http://genome.jgi-psf.org/Agabi_varbur_1/Agabi_varbur_1.home.html. 028-1 homologs were found using BlastP on Uniprot (http://www.uniprot.org/blast/) and PSI-Blast on GenBank.
Alignment between sequences of Ascomycota and Basidiomycota homologs of 028-1 was performed using Cobalt alignment tool (http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi) [21]. Alignment between Basidiomycota homologs and between Yap1 redox domains was done with MUSCLE (http://www.phylogeny.fr/). Alignment between chain A from PAP1, NLS and bZIP was done using BlastP on nr database on these parts of the sequences and BlastP two sequences on smaller fragments. When necessary and available, intron position on the gene was reported on protein sequence to improve alignment.
Domains were determined with CD-search on Conserved Domain Database, CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and with InterProScan Sequence Search (http://www.ebi.ac.uk/Tools/pfa/iprscan/). Putative Nuclear Export Signal (NES) motif position was determined by sequence and position similarity and by determining the presence of hydrophobic amino acids characteristic places [22]. Putative Nuclear Localization Signal (NLS) was localized using NLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) and the homology of this part of the 028-1 sequence with the yeast NLS was determined.
2.6 028-1 Polymorphism analysis
In addition to the 25 wild strains of A. bisporus used in the experiment of mycelium thermo-tolerance, a homokaryon Bs0026-7 [20] from the cultivar hybrid HU1 was used for sequencing. This homokaryon contained the complementary sexual pole of the homokaryon of the hybrid HU1 that had been used at JGI (H97) (DOE Joint Genome Institute) for the whole genome sequencing of A. bisporus var. bisporus. (http://genome.jgi-psf.org/Agabi_varbisH97_2/Agabi_varbisH97_2.home.html). 028-1 sequence identified in the genome of H97 A. bisporus var. bisporus was used for polymorphism studies, as well as the sequence from the genome of A. bisporus var. burnettii, homokaryon JB137-S8 (http://genome.jgi-psf.org/Agabi_varbur_1/Agabi_varbur_1.home.html).
For DNA extraction, stock cultures were made on Petri dishes containing 25 mL of solid CristoMalt agar medium (Barley malt powder, Dif.A1) incubated for 21 days at 25 °C in darkness. The mycelium was harvested, deposited in 2 ml tubes and frozen at −80 °C. Mycelium was lyophilized and stored at 4 °C until DNA extraction and purification.
Genomic DNA was extracted and purified from mycelium using DNA extraction Kit Phytopure (GE Healthcare) following the manufacturer's protocol. PCR primers were designed on 028-1 mRNA sequence (GenBank a.n.: DQ235473) using the Primer 3.0 software v.0.4.0 [23]: ThF (5′-CCCTTCAATAAGCCCTGTCA-3′), ThR (5’-CCTGGATTGCGCTATAATCG-3’). The PCR mixture (50 μL) contained template DNA (30–46 ng μl−1), 2 μL; primer (10 μM), 2 μL of each; dNTP mix (1.2 mM) (Eurobio, France), 5 μL; 5X colorless GoTaq buffer (Promega, Madison, WI), 10 μL; GoTaq DNA Polymerase 2 U (Promega M830B), 0.2 μL; Bovine Serum Albumin (10 mg/ml) (Promega R396D) 0,5 μL. PCRs were run on Eppendorf Mastercycler (USA) with an initial cycle of denaturation at 95 °C for 5 min followed by 34 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1.5 min, and synthesis at 72 °C for 1.5 min with a final extension at 72 °C for 5 min. Amplified DNA (750–850 bp) was separated on a 1% agarose gel in 0.5 × Tris/Borate/EDTA (TBE) buffer to identify the failed reactions. Sequencing of amplified products was performed by Cogenics (Beckman Coulter genomics, United Kingdom). Resulting sequences were aligned using CLUSTAL W software [24]. Some sequences were then subjected to PSI-BLAST [25] for homology search.
Agaricus bisporus var. bisporus has predominantly a pseudo-homothallic life cycle giving rise to heterokaryotic mycelium with potentially two different alleles for 028-1 gene. The sequences of three homokaryons and of two strains suspected to be haploid (A. bisporus var. eurotetrasporus) obtained by sequencing or from the A. bisporus genome were used as reference sequences. Moreover, some strains were homozygotes for the analyzed sequences. In four heterokaryotic mycelia, the presence of at least one deletion in one of the two alleles of the gene and reading in both directions was used to recover the sequence of each homokaryon by progressive manual realignments using BIOEDIT 7 [26]. All these data were used to deduce the two putative haplotypes corresponding to the heteromorphic sequences in the other strains.
Each sequence was tagged with the reference code of the strain and (1) or (2) for the sequence of each of the two nuclei of the heterokaryons. A list of polymorphic sites including indels and nucleotide polymorphisms was generated. The 27 heterokaryons, 2 homokaryotic strains and 3 homokaryons represented 59 haplotypes. The sequences were aligned with ClustalW and analyzed to identify the positions of nucleotide polymorphism.
The phylogenic tree was constructed using neighbor-joining (NJBIO) method [27] implemented in the Seaview program v4 [28]. First sequences were aligned with MUSCLE (v3.7) [29]. Gap sites were include in the NJBIO analyze. Reliability for internal branch was assessed using the bootstrapping method (1000 bootstrap replicates).
3 Results and discussion
3.1 Expression of 028-1 in the vegetative mycelium during heat treatment
Differences in expression levels of 028-1 due to incubation at 32 °C for 12 h were studied in vegetative mycelium of eight strains cultivated at 25 °C. The treatment at this higher temperature induced a significant but relatively low over-expression of 028-1 (Fig. 1) for four out five strains which had been shown to produce fruiting bodies at 25 °C [3]. For Bs0679 which mycelium maintained a low growth rate at 33 °C (Table 1) and two other strains, there was no significant regulation. Bs0261 was slightly affected, but with a down-regulation. The basal level of expression in mycelium of thermo-tolerant strains maintained at 25 °C was not different or 2 to 3 time lower than in susceptible strains.
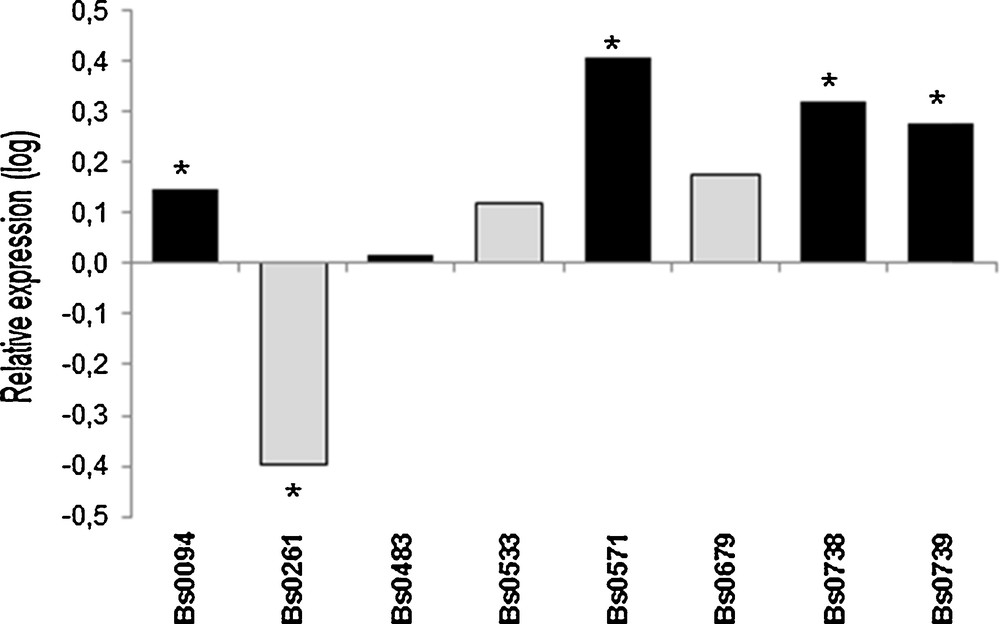
Effect of a treatment for 12 h at 32 °C on vegetative mycelium grown at 25 °C. Relative expression of 028-1 gene (32 °C for 12 h vs. continuous growth at 25 °C). Values are log-transformed data of expression ratios after normalization against the housekeeping gene EF1-α. Significantly different at *P < 0.001. Black bars are FHT+ strains, grey bars are FHT− strains as defined in Largeteau et al. [3].
Chen et al. [14] constructed a binary expression vector of thermo-tolerance-related gene 028-1 of a specific thermo-tolerant strain of A. bisporus and transferred it into a non-thermo-tolerant strain by Agrobacterium-mediated transformation. Most of the 10 transgenic strains cultivated in compost to estimate their colonizing ability at different temperatures showed an obviously increased thermo-tolerance. Our own work with different strains and measurements of transcription levels showed that the up-regulation of 028-1 should be one of the factors associated with the reaction of the vegetative mycelium of some A. bisporus strains to increases in temperature. The involvement of 028-1 in the thermo-tolerance of the vegetative mycelium being confirmed only for some strains, it was worth to characterize this gene and to define its DNA sequence polymorphism in a larger sample of strains.
3.2 Characterization of 028-1 gene and its corresponding protein
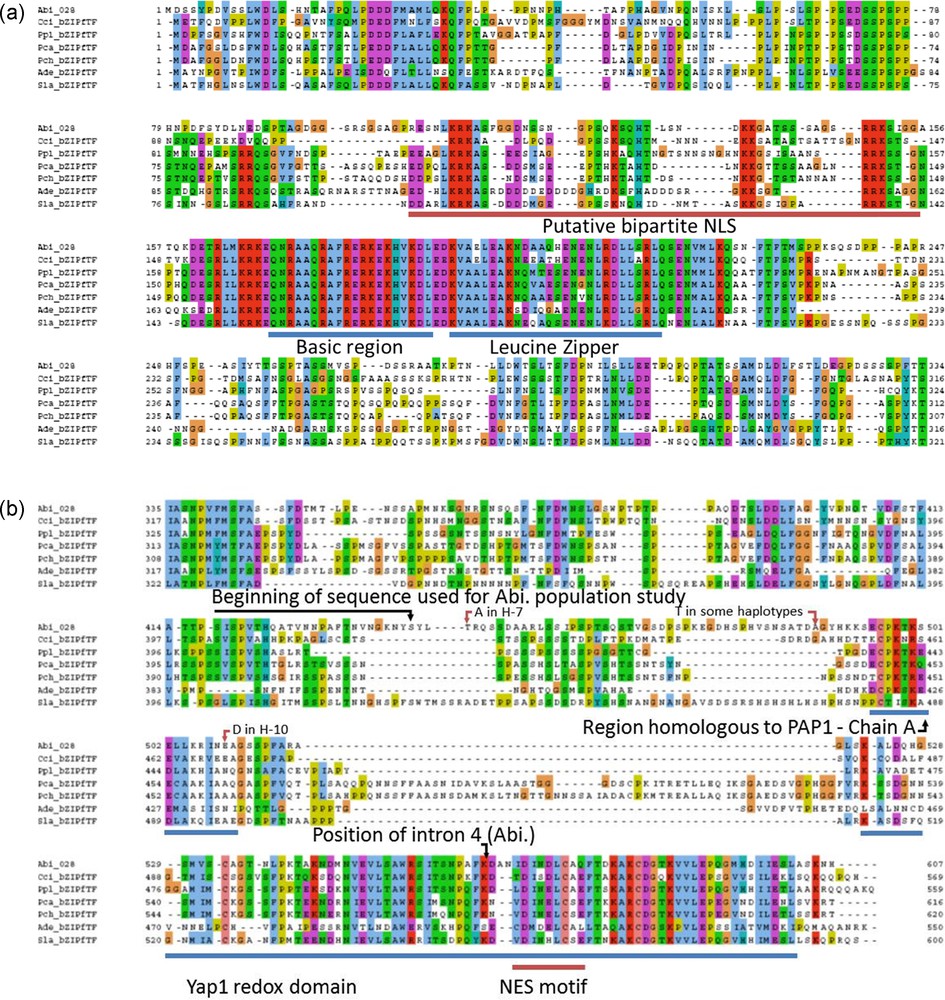
The gDNA sequences of the 028-1 gene from 24 wild strains and 3 homokaryons of A. bisporus were obtained and they allowed completing the information on this thermo-tolerance gene. By alignment of the cDNA sequence from Chen et al. [15] and gDNA obtained here, two exons and one intron of 54 bp were identified. A putative reading frame was defined by identifying ORFs and verifying that the polymorphic bases were mostly in the third place of the codons in exons. Positions of stop codon and of the beginning of 3’UTR were deduced. A new sequence of 028-1 gene was deposited in GenBank under the accession number (a.n.) FJ940724.1. The intron was localized at position 370-423 and a.n. of the corresponding protein sequence is ACR25139.1. The deposited sequence belongs to a wild strain from England being homozygote for this gene fragment. It corresponded to the most frequent allele found in our sample (see below).
This sequence annotation was coherent with models generated by the JGI software on the genome of A. bisporus H97. We improved the automatically generated model by using the homology with, Laccaria bicolor, Coprinopsis cinerea and Phanerochaete chrysosporium homologous sequences and found the beginning of the gene (Fig. 2). This was confirmed by the similarity found on the genome of A. bisporus var. burnettii JB137-s8 (Protein ID 144436). The model generated has the protein ID 239429 on H97 genome v2 and was released in GenBank (EKV52013) with the publication of A. bisporus H97 genome sequence [30]. The transcript and CDS sequences are 2.18 and 1.82 kb respectively, larger than the full-length cDNA sequence published by Chen (DQ235473).
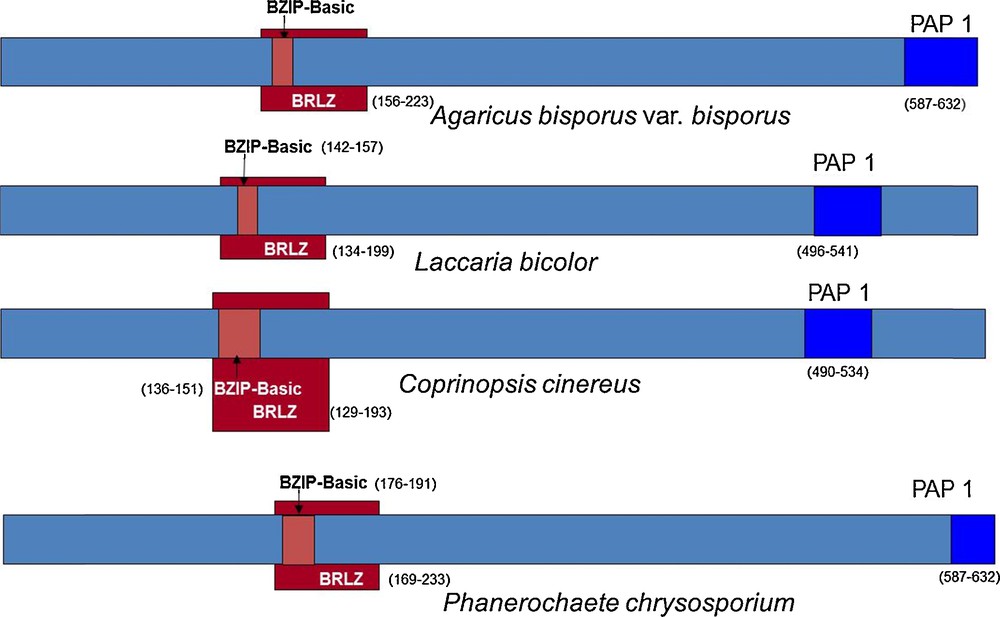
Annotation of the gene 028-1 of A. bisporus var. bisporus and orthologs in Laccaria bicolor (JGI protein ID: 66554), Coprinopsis cinerea (XP_001828489), Phanerochaete chrysosporium (Prot ID 7851). Three domains were first identified with InterProScan software (v4.8). The overall structure is conserved which helped us to find the beginning of the gene. BRLZ - basic region leucin zipper (SMART method; identifier: SM00338). BZIP-Basic (PROSITE method; identifier: PS00036). PAP1 (PFAM method; identifier: PF08601).
The determination of the primary sequence of 028-1 allowed progress in the identification of its role. After performing a research of homology with already identified genes in Search database Reference proteins (ref seq_protein) using BlastP and PSI-BLAST, matches were found with a hypothetical protein CC1G_08635 from C. cinerea: 71% coverage and E-value 9e-32, identities of 51%, and with other Basidiomycota proteins. These sequences were aligned with MUSCLE (Supplementary Fig. S1). A BLASTP in NCBI allowed identifying a homolog in yeasts: AP-1-like transcription factor Yap1 (Yap1p).
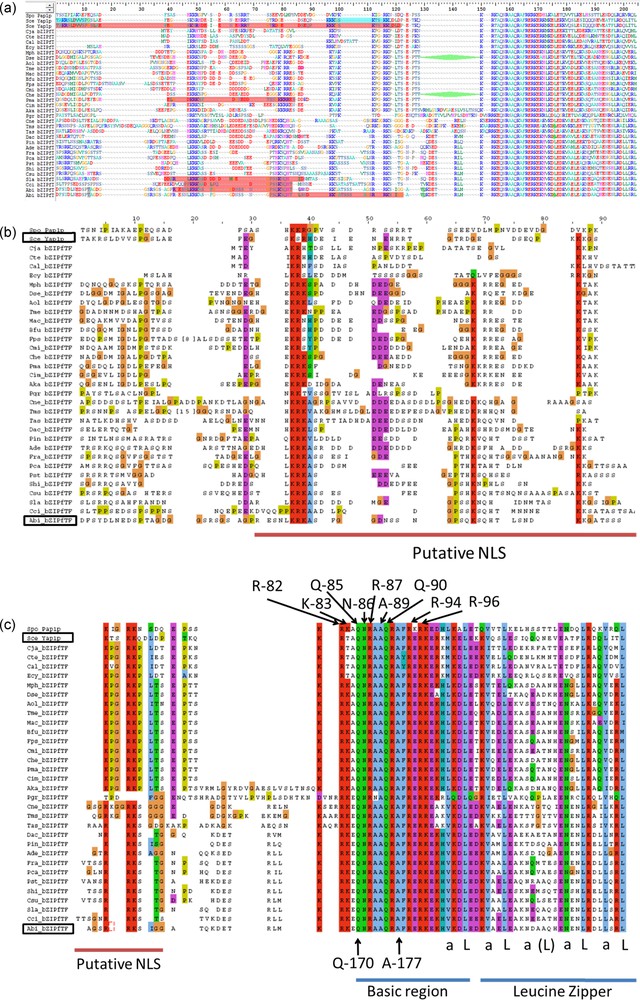
Among Agaricales, the two first proteins containing the Yap1 redox domain to be introduced in UniProtKB database were, the putative uncharacterized protein A8N0T8 from C. cinerea and the A. bisporus protein sequence C4PFX0 (ACR25139.1 in GenBank) from this work. Two domains and two motifs present in Yap1p were identified in A. bisporus 028-1: the bZIP domain with two regions, the Yap1 redox domain, one putative NLS and two putative NES motifs.
The bZIP domain contains a basic region mediating sequence-specific DNA binding followed by a leucine zipper region required for dimerization. PAP1p is an orthologue of Yap1p in Schizosaccharomyces pombe [17] or the Yap1p closest functional homolog, involved in drug resistance and oxidative stress response [31]. The 9 amino acids (R-82, K-83, Q-85, N-86, R-87, A-89, Q-90, R-94 and R-96) engaged in salt bridges or hydrogen bonds with either bases or phosphate groups of DNA in PAP1p/DNA crystallographic complex [31] are all conserved in A. bisporus 028-1 (Fig. 3, Supplementary Fig. S2c). The two S. cerevisiae YAP family-specific residues, a glutamine Q-73 and an alanine A-80, that differ from R-234 and R-241 of the yeast AP-1 transcription factor Gcn4p [17], are conserved and correspond respectively to 028-1 Q-170 and A-177 (Supplementary Fig. S2). Consequently the 028-1 protein is a member of YAP family of AP-1 transcription factors and should have the same DNA binding specificity as PAP1P. The binding sites of YAP family members, YAP Response Element (YRE), are composed of two TTAC “half sites” positioned either in an adjacent (TTACGTAA referred as YRE-A) or in an overlapping fashion (TTA(C/G) TAA referred as YRE-O) [31]. These authors found that a supplementary adenine was present in 5′ of the consensus sequence of many YRE motifs. As PAP1p transcription factor (TF) interacts with both 7 (YRE-O) and 8 (YRE-A) base pair YREs, the same specificity is expected for 028-1 protein. The sequence of the basic region of the bZIP domain is conserved among all Agaricomycetes (Sebacinales, Auriculariales, Polyporales, Corticiales, Russulales, Boletales, Agaricales) and Dacrymycetes putative TF mentioned in Supplementary Fig. S2, the DNA binding specificity of these sequences should be also the same as PAP1P.
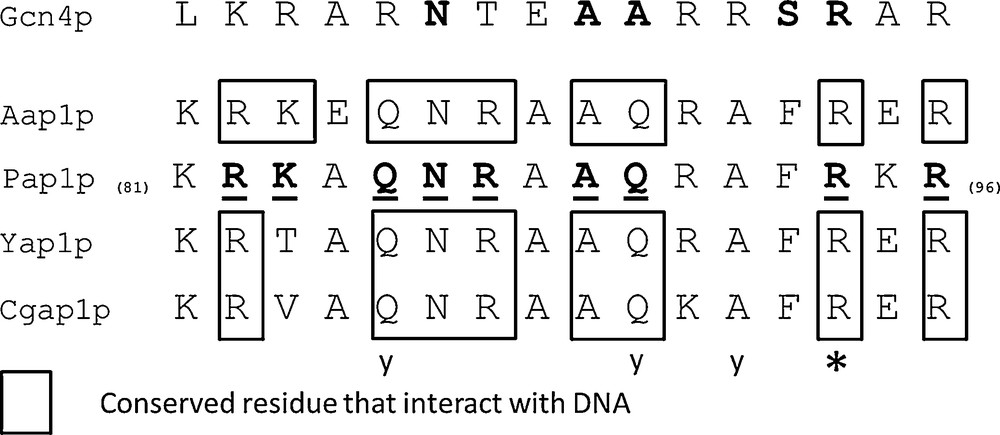
Sequence alignment of the Basic Region (BR) of bZIP domain of AP-1 like transcription factors. Aap1p (028-1 protein) BR is compared with Ascomycetes homologous domains from YAP family transcription factors (PAP1p - Schizosaccharomyces pombe, Yap1p - Saccharomyces cerevisiae, Cgap1p - Candida glabrata) and JUN family (Gcn4p - Saccharomyces cerevisiae). Amino acids underlined interact with DNA in PAP1p/DNA crystallographic complex [31] and are all conserved in A. bisporus Aap1p. Positions in bold in Gcn4p are those common to most bZIP family members. An invariant arginine, at a position labeled with a star, could interact with the central guanine in the Gcn4p/DNA complex (DNA consensus sequence ATGA(C/G)TCAT) [54,55] or in the PAP1p/DNA complex (one of the two DNA consensuses identified TTACGTAA) [31]. Letter y indicates three positions where amino acids are specific to YAP family.
In 028-1 leucine zipper region, the conserved leucines (or other residues) at position d in a helical wheel diagram of the coiled coil and typically hydrophobic residues at position a of the coiled coil are the same as those of Yap1p, or of the two Ascomycota homologs Candida albicans Cap1P or S. pombe PAP1p [17] indicating a typical functional leucine zipper. The two functional regions of bZIP domain are present in A. bisporus 028-1 and well conserved in Basidiomycota homologs.
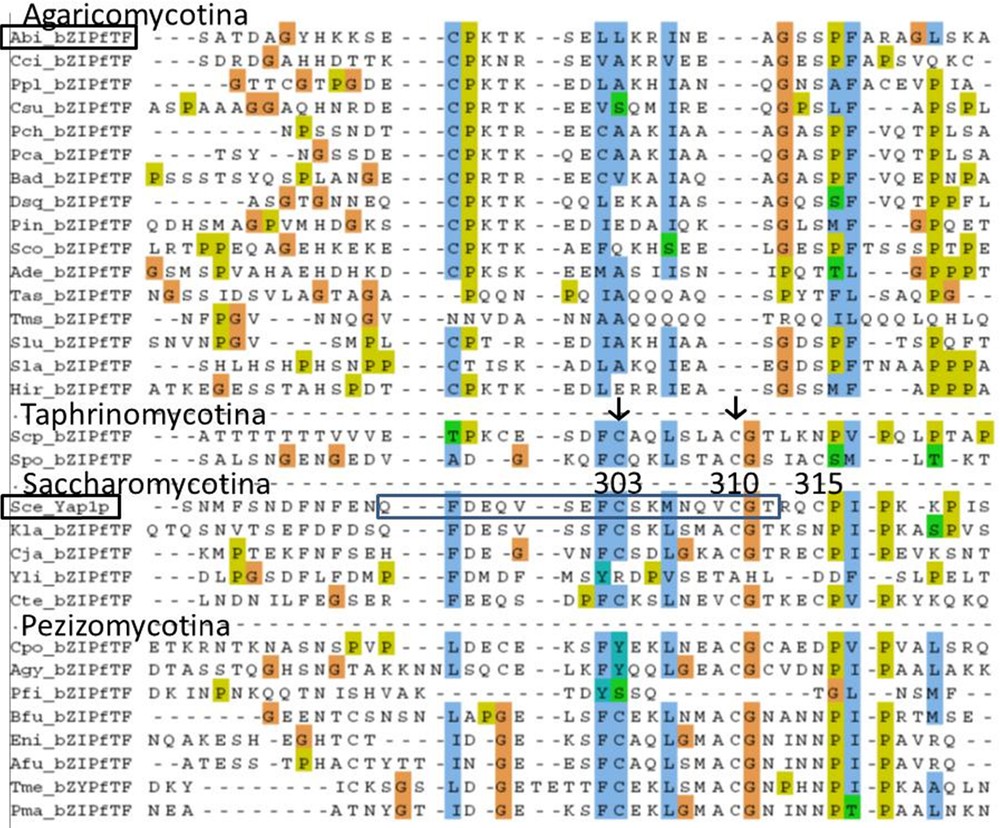
Interproscan in InterPro database allowed identification of the Yap1 redox domain IPR023167 (E-value: 3.7E-23) and the region called Transcription factor PAP1 IPR013910 (E-value: 3e-07) in 028-1 of A. bisporus. PAP1 region was described on PAP1p (AP-1-like transcription factor QO1663 of S. pombe). PAP1 signature is used to build the Pfam family PF08601. The homologous sequences of the chains A and B of PAP1 and YAP1 genes were identified, but the A. bisporus sequence homologous to chain A is rather different. The Yap1 redox domain, correspond to the chain B of transcription factor PAP1.
A homology with chain A of PAP1 (N-terminal Cysteine-riche domain, n-CRD) was difficult to find. By successive pairwise comparison with BlastP we found a putative homology between region PAP1 chain A from Saccharomyces cerevisiae Yap1p DAA09892 (273-323) and selected regions from bZIP fTF of Saitoella complicata (Taphrinomycotina), Schizophyllum commune, A. bisporus 028-1 (Table 2). An alignment between 30 sequences homologous to region PAP1 chain A of S. cerevisiae in Dicarya was obtained (Supplementary Fig. S3). The most conserved part of chain A in PAP1 region in Saccharomycotina (pfam08601) correspond to Yap1 sequence 294-QFDEQVSEFCSKMNQVCGT-312 that includes the two cysteines involved in disulfide bonds present in the oxidized form of Yap1p (Cys303-Cys598 and Cys310-Cys629) [32]. The alignment in Supplementary Fig. S3 indicates that these two Cysteine positions (Cys303 and Cys310) are conserved in many (but not all) Saccharomycotina, Taphrinomycotina and Pezizomycotina. Therefore the redox sensing mechanism involving the disulfide bonds formation is probably present for all these Ascomycota Yap1p homologs (with chain A two Cys positions conserved) and this mechanism was probably present in the ancestor of all Ascomycetes. This part of chain A is homologous to 028-1 sequence 495 - ECPKTKSELLKRINEA - 510. The alignment indicated no homology between Cysteines of chain A from Sce_Yap1p (C-303, C-310) and those of 028-1. None of these disulfide bonds existed in A. bisporus, neither in other Basiodiomycota bZIP fTF studied. The redox sensing process in Basidiomycota bZIP fTF studied involves another mechanism than that described in S. cerevisiae.
Inferred homologies for Chain A of Transcription factor Pap1 region between Saccharomycotina (Saccharomyces cerevisiae), Taphriomycotina (Saitoella complicata), Agaricomycotina (Schizophyllum commune, Agaricus bisporus) and between Saccharomycotina (S. cerevisiae) and Pezizomycotina (Tuber melanosporum). For phylogenetic tree of Dicarya, see http://genome.jgi-psf.org/programs/fungi/index.jsf.
| Compared putative chain A sequences | Accession number of protein complete sequence | Identity | Similarity | Number of positions | Blastp two sequences result | |||||||
| S. cerevisiae Yap1p | DAA09892 | Sbjct | 18 | SEFCSKMNQVCGTRQCPIPKKPIS | 41 | |||||||
| 0.38 | 0.75 | 24 | S+FC++++ CGT + P+P+ P + | |||||||||
| S. complicata bZIPfTF | 15991 | Query | 8 | SDFCAQLSLACGTLKNPVPQLPTA | 31 | |||||||
| S. complicata bZIPfTF | 15991 | Query | 4 | PKCESDF | 10 | Query | 22 | KNPV | 25 | |||
| 0.36 | 0.82 | 11 | PK +++F | ++P | ||||||||
| S. commune bZIPfTF | EFI91622 | Sbjct | 17 | PKTKAEF | 23 | Sbjct | 32 | ESPF | 35 | |||
| S. commune bZIPfTF | EFI91622 | Query | 7 | QAGEHKEKECPKTKAEFQKHSEELGESPFT | 36 | |||||||
| 0.57 | 0.63 | 30 | AG HK+ ECPKTK+E K E G SPF | |||||||||
| A. bisporus bZIPfTF (028-1) | EKV52013 | Sbjct | 4 | DAGYHKKSECPKTKSELLKRINEAGSSPFA | 33 | |||||||
| S. cerevisiae Yap1p | DAA09892 | Query | 21 | NQFD-EQVSEFCSKMNQVCGTRQCPIPK | 47 | |||||||
| 0.39 | 0.57 | 28 | D E + FC K++ CG PIPK | |||||||||
| T. melanosporum bZIPfTF | XP_002837914 | Sbjct | 28 | GSLDGETETTFCEKLSMACGNPHNPIPK | 55 |
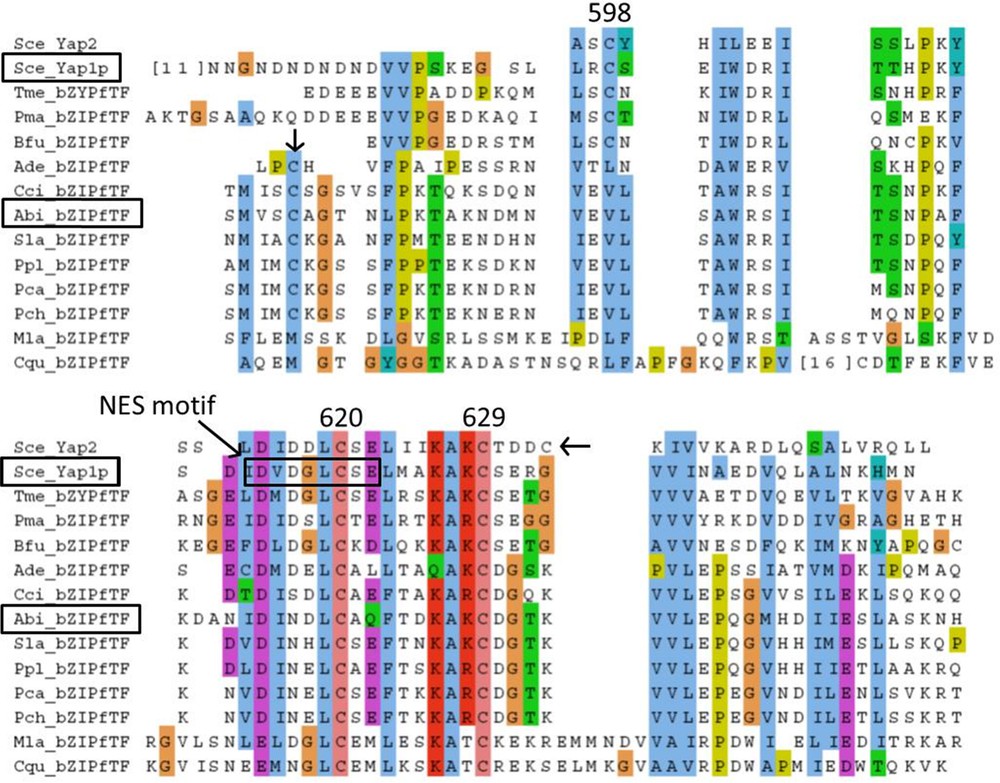
The Yap1 redox domain, correspond to the chain B of transcription factor PAP1. The middle of this C-terminal Cysteine-riche domain (c-CRD) is very similar between Ascomycota (including Yap1p) and 028-1, and position of Yap1p Cysteine C-629 is conserved in 028-1. Nevertheless the region surrounding the Cysteine on the C-terminal side of Yap1 redox domain in Yap1p is not homologous to the corresponding region of 028-1 (Supplementary Fig. S4).
The Yap1p, Nuclear Export Signal (NES) motif is nested in the Yap1 redox domain. It contains hydrophobic residues and several acidic residues in between, at positions conserved in Ascomycota homologs (Supplementary Fig. S4). Most of these characteristics are shared with viral and metazoan NES [22] and are conserved at the same position in Yap1 redox domain of 028-1 protein. Therefore we can deduce the presence of a putative NES signal in 028-1 protein at position 568-576. Another putative NES motif was found on 028-1 protein using NetNES1.1 Server (http://www.cbs.dtu.dk/services/NetNES/) at position 313-324. In yeast an exportin, Crm1p, recognizes the NES of the reduced form of Yap1P and directs it out of the nucleus. We have found a homolog of Crm1p among the gene models of the genome of A. bisporus [30], GenBank accession number EKV50484. This gene has 52% identities and 74% similarities with yeast Crm1p (a.n. DAA08312). It contains an Xpo1 domain, pfam08389 (E_value 2.41 e-46) and an Exportin 1 domain also known as CRM1_C-terminal domain, smart01102, (E_value 2.15 e-151). Unlike many member of the family pfam08389, the Xpo1 domain of A. bisporus Crm1p-like is exactly at the same position than in yeast exportin 1 (Crm1p) that is close to the N-terminus. A. bisporus Crm1p-like is a putative exportin 1 homolog that could link to the NES motif of 028-1. The mechanism by which Yap2p and Yap8p, the two other yeast proteins having a Yap1 redox domain, and Yap1p react to Cadmium, Arsenic concentration and thiol reactive agent respectively don’t involve formation of disulfide bonds [19,33]. Experiment is needed to control to which stimulus 028-1 protein is able to respond and to determine the mechanism involved.
Next to the BZIP domain of Yap1p a NLS motif enables Pse1p, to bind the protein and to transport it in the nucleus through the nuclear pore complex [34]. Pse1p is an importin beta family member. Using cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) we found a possible location for a putative bipartite importin alpha-dependent nuclear localization signal (NLS) on 028-1 protein sequence near the N-terminal side of the BZIP domain: RESNLKRKASFGGDNSSNGPSQKSQHT with a score 5.6 (Supplementary Fig. S2a). Another location, for a putative bipartite NLS, was found with cNLS Mapper on 028-1 protein, overlapping the first one, but with a smaller score: PSQKSQHTLSNDKKGATSSSAGSRRKSIGG with a score 4.
As indicated in Supplementary Fig. S2 the cNLS Mapper prediction and the experimental NLS localization on Yap1p were slightly different. Moreover, the NLS of Yap1p and Pho4p, another target protein of Pse1 in yeast, did not fit exactly to the consensus basic patterns of the classic NLSs. Such kind of differences between their model and experimental data were also observed by Kosugi et al. [35] when they designed their software. Analysis of this is given in Supplementary Fig. S2. By combining cNLS mapper results and sequence alignment we predicted that 028-1 bipartite NLS could be inside the region 107-155, with one N-terminal region including the sequence LKRKAS and one C-terminal region including the RRK sequence.
The gene models of the genome of A. bisporus [30], contain a homolog of Pse1p, GenBank accession number EKV48756. This gene has 25% identities and 44% similarities with yeast Pse1p (a.n. EIW08583). It contains an Armadillo/beta-catenin-like repeats (ARM domain), cd00020 (E_value 2.39 e-06) and two Heat-like repeats (domain HEAT_EZ) pfam13513 (E_value 6.26e-04 and 3.71e-03). These domains are present and well conserved on the N-terminal side of these two proteins, but there is only one HEAT_EZ domain identified on Pse1p as the second one diverged a little more from the consensus than in Abi_ EKV48756. The C-terminal side of Pse1p is 213 amino acids longer than Abi_EKV48756 which is a putative importin beta family member that could link to the NLS motif of 028-1 and we named it Pse1p-like protein.
As a summary, except for domain PAP1 chain A, for which the Cysteine positions are not conserved, all the domains and motifs of Yap1 can be predict on 028-1 protein sequence. Sequence similarities as well as domain and motif predictions indicated it has the structure of a TF with a sequence-specific binding domain and a leucin zipper region. It has a potential sensing capacity with a Yap1 Redox domain and it should be able to transfer in and out the nucleus thanks to the NLS and NES motifs.
Among the 15 A. bisporus genes uncoding proteins containing a BZIP-basic domain (IPR004827), 028-1 is the only one uncoding a Yap1 redox domain or a partial PAP1 region (PF08601). Then 028-1 is the Yap1p homolog and could play a similar role in A. bisporus than in the yeast. The transcription factor (TF) Yap1p is a central regulator of the response to oxidative stress in S. cerevisiae [17,19]. In presence of H2O2, reactive oxygen species (ROS) and Cd, cellular localization of Yap1p is modified and this TF induces expression of genes involved in detoxification of ROS, thiol redox control, production of antioxidants and also cadmium detoxification pathways. However, since a putative common ancestor to YAP1 and 028-1, the gene could have evolved differently in Basidiomycota and Ascomycota.
We annotated 028-1 homologs in available genomes of Basidiomycota showing the conservation of the protein domains in their structure (Supplementary Figs. S1, S2, S4). Alignment of gene sequences showed the high level of identity in the domains B_ZIP1 and Yap1 redox and also, at a lesser level in the sequence located between the two domains and before B_ZIP1 (Supplementary Fig. S1). However an indel of 42 to 48 amino acids, between n-CRD and Yap1 redox domain, distinguished Phlebioid clade (Phanerochaete) of Polyporales from Antrodia clade and all other Basidiomycota tested. The important polymorphism found in this gene in Polyporales could be used to control or improve Polyporales phylogeny which is not stabilized yet [36,37]. The high level of identity between Basidiomycota protein tested here indicated they are potentially orthologous proteins. However we have not been able to align the sequence located between the two domains when comparing Basidiomycota with Ascomycota and had great difficulty to find the homologous sequence of PAP1 chain A in Basidiomycota. Therefore the similarities between the inferred structures of 028-1 and Yap1p proteins can be confirmed, without ambiguity, only at the level of the two domains and the NES motif. We demonstrated that the NLS motif has evolved much more in its position and sequence, with an important indel distinguishing Ascomycota from Basidiomycota homologs. All these evolutions may have modified potential interactions with Pse1p-like proteins. All these differences, between Ascomycota Yap1p homologs and Basidiomycota 028-1 protein homologs, indicate that the function of these proteins could have slightly diverged since the ancestor protein. Indeed we have demonstrated the absence of homology between Ascomycota and Basidiomycota studied, for some Cysteines involved in disulfide bond in oxidized form of Yap1p. Moreover Goudot et al. [31] comparing transcriptional modules (TMs = set of genes whose transcription is modulated by a common transcription factor) of three yeast AP-1 proteins (S. cerevisiae Yap1p, Candida glabrata Cgap1p and C. albicans Cap1p) showed important rewiring in their TMs while their physiological role in response to oxidative stress was conserved.
This survey shows that 028-1 is related to transcription factors activated during stress response in fungi. This homology is not surprising since 028-1 was identified after a temperature treatment on mycelium, and the effect of such treatments on its expression level has been confirmed for some strains in the present work. 028-1 protein is the A. bisporus protein most similar to Yap1p. Even if 028-1 protein does not regulate export from the nucleus by exactly the same mechanism as Yap1p, it is conceivable that it could play a role in oxidative stress response. Sugiyama et al. [38] demonstrated a link between heat shock response in yeast and Yap1p-dependant induction of glutathione synthesis. Heat shock stress increases oxygen respiration rate and intracellular oxidation level and Yap1p senses this increase of oxidation level [39] and activates transcription of its target genes.
Based on these observations, 028-1 gene in A. bisporus may be considered as a transcription factor involved in regulation of the response to oxidative stress generated by heat shock. It might consequently act during the adaptation phase leading to the thermo-tolerance of some strains. We have named it aap1 for Agaricus bisporus activating protein Yap1-like transcription factor 1 as it is a member of a new subfamily of Yap1-like transcription factors, present in Agaricomycetes and Dacrymycetes and not able to form disulfide bonds in the same way as Yap1p in oxidized environment.
3.3 Sequence polymorphism of aap1 in A. bisporus strains
Differences in promoter or coding sequences might, however, be involved in the variability of phenotypes observed in the 24 wild strains of A. bisporus chosen to represent the three known varieties of this mushroom, var. bisporus, var. burnettii and var. eurotetrasporus, and originating from various European and North American sites (Table 1). Three sequences from homokaryons were added as controls to this sample. A. bisporus is an amphithallic species with a secondarily homothallic or heterothallic cycle according to the ploïdy level of the spores which can be heterokaryotic (n + n) or homokaryotic (n), respectively. The vegetative mycelium obtained after isolation from the cap is heterokaryotic, except for the homokaryotic variety A. bisporus var. eurotetrasporus [1,2]. Consequently, a maximum of 59 haplotypes were expected from the studied sample containing 27 heterokaryons, 2 homokaryotic strains and 3 homokaryons. Among them, 54 were identified (Table 1).
Analysis of DNA sequence polymorphism in a gene may be used in molecular ecology research, including gene mapping [40]. In the 701 bp segments of 028-1 gene analyzed, (GenBank a.n.: FJ940724; position 1 corresponding to position 2097 of total gene in JGIDB: Agabi_varbisH97_2_239429), at least 16 nucleic different haplotypes were present (Table 1) with 42 polymorphic sites showing the relatively high level of polymorphism of this gene.
Ninety-three percent of these polymorphisms were single-base changes of which 72% were transitions and 28% transversions. Two percent were three bases possible for the same site. The other polymorphisms (5%) were 2 insertion-deletion polymorphisms (indels) located in the intron. Väli et al. [41] introduced the use of short indels for genetic analysis of natural populations and stated that indels will form an important source of genetic markers, easy and inexpensive to genotype, for studies of natural populations. Active transposons are believed to be the causes of large Indel polymorphisms in maize [42]. Insertions and deletions can occur by unequal crossing-over or replication slippage or slipped-strand mispairing [43]. Here a haplotype differing by an indel was found in two European strains (H-11 in France and Greece), another haplotype differing by another indel was found in two North American strains (H-7 in Canada and USA), independently of their adaptation to high temperature.
Wang et al. [44] stated that Cytosine residues within CpG dinucleotides are the most mutable sites within the human genome, because most are methylated and can spontaneously deaminate to yield a thymidine residue. Our observations agreed with this model stating a frequent occurrence of the reactions of deamination of the cytosines [45]. 32.5% of the single-base changes (SNP) occurred at dinucleotides CpG, and C↔T substitution where 25% of these SNP, whereas they make up about 13% of the sequence.
Of 27 heterokaryotic strains, at least two (Bs0374 and Bs0679) were homozygotes for the studied part of 028-1 gene. The polymorphism at position 117 and 186 allowed distinguishing some A. bisporus var. burnettii haplotypes (T for H-8, H-9 and H-10 at position 117 and A for H-8 at position 186) to the haplotypes of the two other varieties. Ten positions (96, 117, 186, 204, 249, 282, 377, 469, 484 and 644) presented a nucleotide only found in at least one nucleus of a strain of A. bisporus var. burnettii (H-8, H-9 or H-10) and never present in the strains of the other varieties. Only one genotype (H-4) was observed for the two strains of A. bisporus var. eurotetrasporus that was also present and frequent in the variety bisporus. This genotype was found also in one of the nuclei of the intervarietal hybrid (var. eurotetrasporus x var. bisporus), the wild strain Bs0261.
Analysis of DNA sequence polymorphism in a gene may also contribute to identify nucleotide regions that could be responsible of differences in the gene regulation or properties and to progress in population and evolutionary studies [46–48]. Mutations observed as single nucleotide polymorphisms are abundant and widespread in many species’ genomes (coding and non-coding regions), and they evolve in a manner well described by simple mutation models, such as the infinite sites model [46]. The relationship between the A. bisporus strains, varieties bisporus, burnettii and eurotetrasporus studied in this work was analyzed by generating useful distance matrixes as input to cluster the homokaryons of the different strains (Table 1) on the basis of their similarity, using the BIO neighbor-joining method (Fig. 4). The two strains and the homokaryon coming from an isolated population in the Californian Desert and belonging to the burnettii variety presented some variability at the level of nucleic sequences with a minor allele each (H-8, H-9 or H-10). The two heterokaryons from this variety are able to produce mature fruiting bodies at 25 °C (FHT+ strains). If aap1 is responsible for the polymorphism of FHT trait, alleles H-8, H-9 and H-10 could be responsible of FHT+ phenotype as they are never found in FHT− strains.
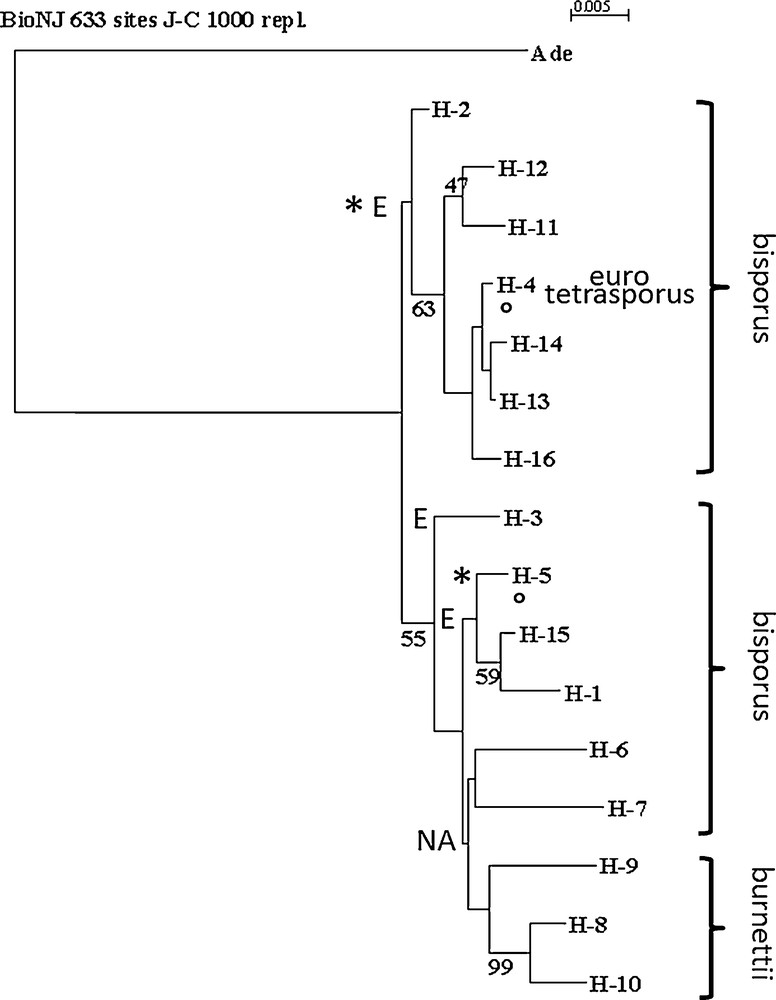
Neighbor-joining tree of aap1 gene haplotypes of Agaricus bisporus strains, obtained by BIONJ analysis using Jukes and Cantor (1969) distance. The tree was rooted by Agaricus devoniensis homologous sequence (Ade). Bootstrap values above 45% (1000 replicates) are displayed beside nodes of the tree formed by, H- 1 to H- 16, A. bisporus haplotypes (Table 1). Varieties to which belong the stains are indicated (bisporus, eurotetrasporus, burnettii). Origin of strains, European (E) or North American (NA) continent, is mentioned. H-1 is one of the most frequent alleles in Europe but has been found in one Mexican strain (Bs0739) below H-4 and H-5 indicate that one stain, Bs0004, having these haplotypes, do not come from Europe but from Canada. This strain or the two aap1 alleles it contains are probably escaped from culture. * beside the European eurotetrasporus bisporus clade and beside H-5, indicate that all these European strains have a threonine at position 47 while all the North American and three other European strains (H-1, H-3 and H-15) have an alanine at this position (position 1 in GenBank a.n. FJ940724 corresponding to position 2097 of total gene in JGIDB: Agabi_varbisH97_2_239429). Position of the root sequence (Ade: Agaricus devoniensis bZIP fTF) is not strongly supported. Maximum likelihood method (PHYML) places, in some cases, Ade near H-16.
In the variety bisporus, Bs0190 and Bs0661 shared two alleles (H-1, H-4), had the same phenotype (Table 1) but had different expression profiles when comparing primordia and fruiting bodies at 17 and 25 °C (see below). The haplotypes from the strains able to produce mature fruiting bodies at 25 °C (FHT+ strains) were distributed in various branches of the tree with the haplotypes from the FHT− strains. However, two main branches were observed in the tree, but with a low bootstrap value. Two haplotypes, H1 and H4, were the most abundant; 27 and 31% of the 55 haplotypes identified respectively. H1 appeared to be neutral for the FHT trait, whereas H4 was less represented in FHT+ (frequency of 0.18) than in FHT− strains (frequency of 0.38). Fisher's Exact Test [49] for count data showed that these differences were significant with a P-value of 0.95. These observations tend to highlight a contribution of some alleles of aap1 gene to the regulation of the fructification by the temperature.
In addition, H4 is present only in European strains, but a haplotype of a strain from British Columbia Canada, whereas all the alleles from the American strains are in the lower branch of the tree. Isolates from British Columbia had already been observed genotypically similar to those collected in Europe [50]. In contrast, the population from forests of Picea in the Rocky Mountains of Alberta from where came the other Canadian strain of this study (a FHT+ strain) had genotypes that were very different from European and cultivar groups [51]. Some alleles, at specific positions in the protein sequence, are specific to North American or European continent (Fig. 5). The DNA sequence polymorphism of 028-1 might be a marker of population evolutions but not an efficient marker of a specific adaptation of thermo-tolerant strains that have been found spread in various populations of A. bisporus var. bisporus.
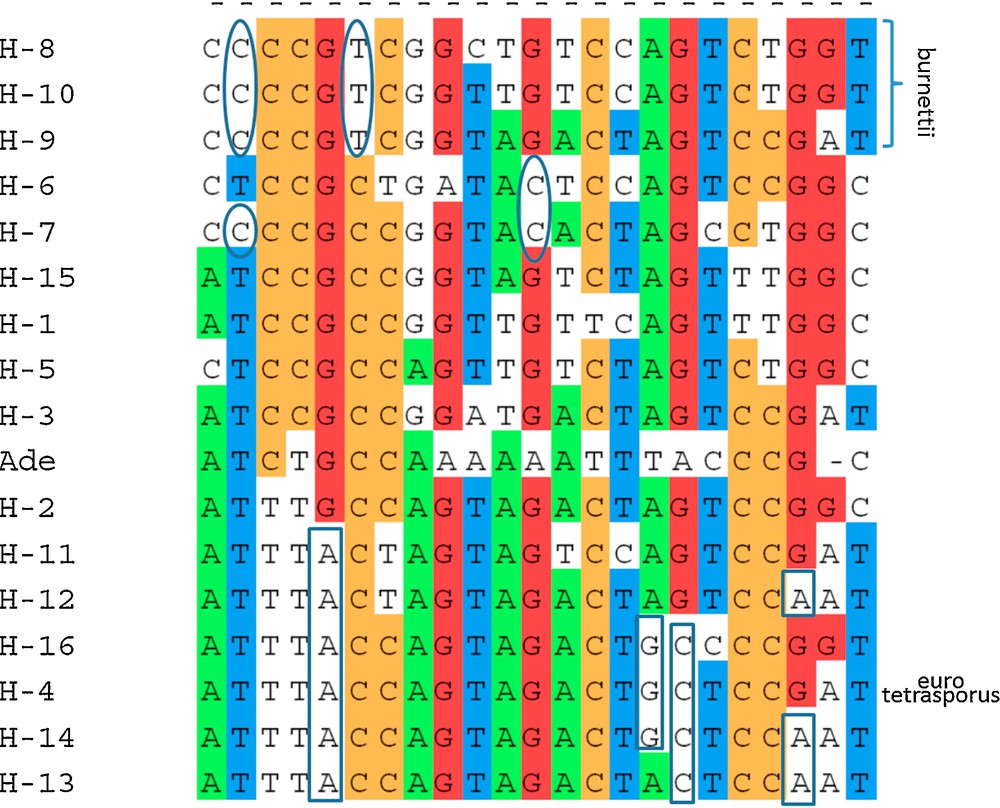
(Color online). Single nucleotide polymorphisms for parsimony: polymorphic nucleotide positions with at least two copies of each allele among the 16 haplotypes of aap1 gene, H- 1 to H- 16 and the root sequence (Ade: Agaricus devoniensis BZIP fTF) used to build the dendrogram (Fig. 4). Positions in gDNA sequence (GenBank accession number FJ940724): 9; 21; 30; 48; 57; 117; 123; 139; 219; 249; 270; 303; 321; 354; 380; 384; 429; 432; 447; 474; 477; 583; 611. Intron position: 370–423. Ellipses: alleles specific to North American strains. Rectangle boxes: specific alleles to European strains but strain Bs0004 from Canada. This strain or the two aap1 alleles it contains are probably escaped from culture. * position 139, SNP on the first base of the codon, dividing the population into two groups one with an adenine, the corresponding amino acid being a threonine (T- 47), the other with a guanine, the corresponding a.a. being an alanine (A-47), cf. Fig. 4. Haplotype H-4 is present in eurotetrasporus and bisporus strains. Three haplotypes belong to var. burnettii, all other haplotypes belong to var. bisporus.
Due to the significant number polymorphic sites in the coding regions, one can question their functionality. Most of the nucleotide changes in the protein-coding regions were silent; they did not change the amino acid composition of the protein product and have generally been assumed to exert no discernible effect on gene function or phenotype [52]. Apart from these synonymous polymorphisms, we identified 3 variable amino acids positions (T/A-4, T/A-47 and E/D-68), of the deduced polypeptide sequence, leading to four different profiles: P1 = TAE, P2 = TTE, P3 = AAE, P4 = TAD. Alignment of the deduced polypeptide sequences from the DNA coding sequences of the nucleus arranged according to the different A. bisporus strains leaded to four groups of phenotypes for the protein Aap1 due to the combination of alleles in the various heterokaryotic strains (Table 3). No specific allele was observed for the mycelium surviving at 33 °C. Comparison between the protein phenotype and the ability of the strains to produce fruiting bodies at 25 °C (FHT+) showed that the profile P1 with amino acids TAE was present in at least one nucleus of 11 out 14 FHT+ strains whatever their origin, whereas 8 out of the 13 FHT− strains have this peptide sequence. Fisher's Exact Test [49] for count data shows that these differences were not significant. Despite the polymorphism of aap1 is not clearly correlated with the thermo-tolerance of the strains, the data does not close the door to a contribution to the expression of the ability of A. bisporus strains to fruit at 25 °C.
Classification of strains based on their Aap1 peptide sequences and their thermo-tolerance.
| Pairs of peptide sequences | Phenotypes | ||
| Fructification at 25 °C | Mycelium survival at 33 °C | Others | |
| 1/1 | Bs0094, Bs0374, Bs0739 | Bs0679, Bs0739 | Bs0420 |
| 1/2 | Bs0190, Bs0419B, Bs0571 Bs0431C, Bs0474, Bs0661 | Bs0085A, Bs0370, Bs0416D, Bs0261, Bs0026 (= HU1) | |
| 1/3 | Bs0483 | Bs0717 | |
| 1/4 | Bs0738 | ||
| 2/2 | Bs0285, Bs0584, Bs0705 | Bs0564 | Bs0004, Bs0572, Bs0423, Bs0514 |
3.4 Expression of aap1 in the thermo-tolerant strains at the fruiting stage
The putative involvement of aap1 in the thermo-tolerance during the first stage of fruiting was studied by measuring the relative expression levels in primordia produced at 17 °C and 25 °C, using a sample of 9 strains selected for their ability to produce mature fruiting bodies at 25 °C [3]. The differences were weak but significant for 4 of the strains. A higher expression was recorded in primordia obtained at 25 °C than in those obtained at 17 °C for Bs0094 and Bs0661 whereas aap1 was down-regulated for Bs483 and Bs738 (Fig. 6). The low levels of regulation and the absence of a general pattern tend to indicate that 028-1 is not involved directly in primordia susceptibility to different temperatures. Previous development stages such as mycelial cords or pinheads should have been more appropriate to unravel a difference in expression of aap1 at 17 and 25 °C.
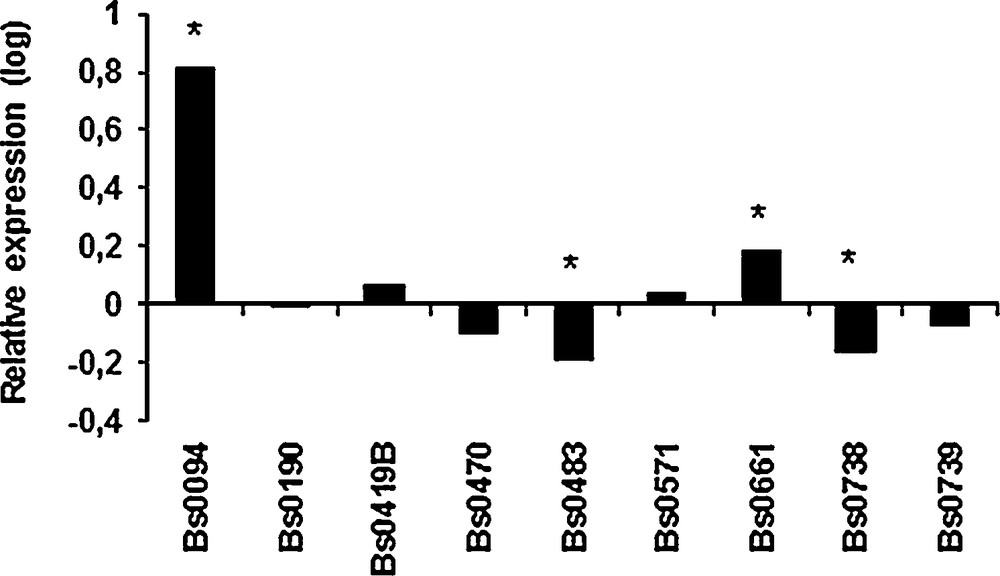
Expression of aap1 gene in primordia obtained at 25 °C relative to expression in primordia obtained at 17 °C for 9 strains of A. bisporus. Values are log-transformed data of expression ratios after normalization against the housekeeping gene ef1-α. Significantly different at *P < 0.001.
Temperature may affect the differentiation of primordia to mature fruiting bodies. We frequently observed that for some strains primordia were produced at 25 °C, but they never gave rise to fruiting bodies. When the expression of aap1 was compared in fruiting bodies and primordia obtained at 17 °C, only Bs0094 showed a significant over-expression in the fruiting bodies, but at very low level (Fig. 7). At 25 °C the gene was up-regulated in fruiting bodies for 3 strains whereas it was down-regulated for Bs0094. Such differences between the strains could be linked to the level of adaptation for fruiting at 25 °C. Moreover, in absence of significant regulation, differences in basal expression levels in primordia could contribute to the involvement of aap1 to their ability to develop into fruiting bodies at 25 °C. It would be interesting to measure basal levels of expression in a large set of primordia obtained at 17 °C from both thermo-tolerant and susceptible strains unable to produce fruiting bodies at 25 °C for comparing the two groups.
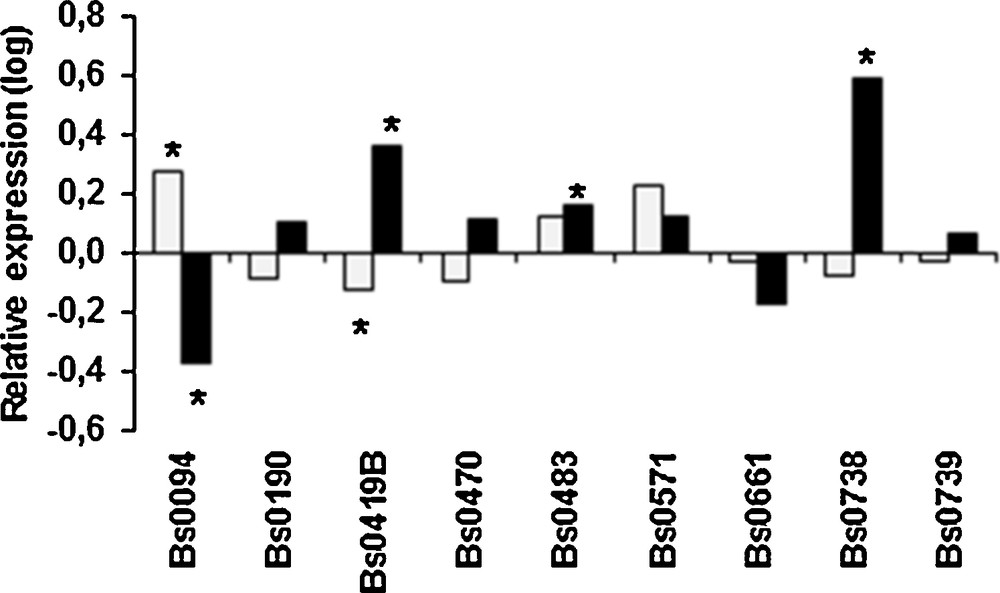
Relative expression of aap1 gene in fruiting bodies vs. primordia obtained at 17 °C (grey) or at 25 °C (black). Values are log-transformed data of expression ratios after normalization against the housekeeping gene ef1 α. Significantly different at *P < 0.001.
The low level and low significance of differential regulation between the cultures at 17 and 25 °C at the two stages of morphogenesis studied here lead to the conclusion that aap1 was not directly involved in the ability of A. bisporus strains to produce mature fruiting bodies at a higher temperature than that used for the cultivated strains.
3.5 Alternative transcriptional initiation hypothesis
Using a rapid amplification of cDNA ends (RACE), Chen et al. [13] obtained a cDNA sequence that begins at position 810 of aap1 coding sequence (ccgaTtctt@810) corresponding to the third base of the codon of aspartate D-270. They suggested that the first methionine found by translating cDNA on the three reading frames, could be the beginning of the translation. This methionine is M-313 in the entire protein sequence and then corresponds to the same reading frame as the whole protein. In the A. bisporus H97 genome portal [30] many EST of aap1 gene (protein ID 239429) are indicated before the beginning of the cDNA found by Chen et al. [13] and no EST begins at the same place as that of Chen et al. [13]. However thermal stress was not used when RNAs were obtained and sequenced to annotate the genome. An alternative transcriptional initiation, from nt 810 of the cds, could occur during stress conditions. The resulting protein would lack bZIP region and NLS motif but not NES motif. It could interact in the cytoplasm with exportin binding the NES motif. Chen and co-workers [14] transformed a non-thermo-tolerant strain with a plasmid vector containing only a partial part of the aap1 gene: their cDNA (028-1). Differential transcriptional initiation of the same gene depending of the thermic conditions could modulate the phenotype. This hypothesis should be tested.
4 Conclusions
This work sets up the first in-depth study aimed to validate the expression of a gene in high temperature fruiting. Unlike the Chinese strain studied by Chen et al. [13–15], the 028-1 gene did not seem to be involved as a major factor in thermo-tolerance of European and American strains of A. bisporus var. bisporus. The sequence of the protein encoded by this gene was established and was identified as an activating protein Yap1-like transcription factor (Aap1). Many Ascomycota and Basidiomycota homologs of aap1 were found. We demonstrated that the redox sensing mechanism involved in Yap1 transcription factor participating to yeast thermo-tolerance was not entirely conserved in this Yap1 homolog. In fact, cysteine positions involved in disulfide bonds were not conserved. That is the reason why we give a new name, aap1, to this gene belonging to a new Agaricomycotina (Basidiomycota) subfamily of YAP1 homologs. Aap1 might be involved in general stress resistance mechanisms and act not directly and only partially in the ability of A. bisporus strains to produce mature fruiting bodies at a higher temperature than that used for the cultivated strains, but it acts indirectly in the vegetative mycelium. Its DNA sequence polymorphism and deduced peptide sequences were not strictly related to the phenotypes but to the varieties and the populations from which the strains were originated. This work highlights the need to study in A. bisporus populations the polymorphism of the sequence susceptible to be the promoter of the gene and of the NLS motif in order to conclude about the putative link between this gene, the FHT trait and the mycelium thermo-tolerance. Two other hypotheses should be tested: (i) are there differences in basal expression levels of the gene, in a large set of strains, at the mycelium or at the primordia stages? (ii) is there a differential transcriptional initiation of the gene depending of the thermic conditions? We also noticed the high variability of the NLS region in Yap1 transcription factor and Aap1 homologs among Dicarya. This variability could play an important role in the unequal ability of these proteins to enter the nucleus to activate the target genes. The recent release of the whole genome sequence of A. bisporus [30] and the publication of an expanded genetic linkage map of an intervarietal Agaricus bisporus var. bisporus and A. bisporus var. burnettii hybrid [53] open new opportunities to search for candidate genes through without a priori approaches.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
Pilar Navarro was supported by a fellowship from CONACYT, Mexico. This work was partly funded by the ECOS-Nord-ANUIS committee, Action M06A01. We thank Charlène Descorps for her contribution in gene expression studies.
Appendix A Supplementary data
a and b. Sequences alignment of Basidiomycota proteins encoded by 028-1 gene homologs. The two domains, B_ZIP1 (basic region + leucine zipper) and Yap1 redox domain (PAP1 chain B) are localized, based on NCBI Conserved Domain database (A. bisporus B_ZIP1 E-value = 8.37e-06, P. carnosa PAP1 E-value = 3.66e-04). A putative bipartite NLS motif was localized using cNLS mapper and by alignment with Yap1 (cf. Fig. S2). A NES motif was deduced by alignment with Yap1 (cf. Fig. S4). An indel of 42 to 48 amino acids, just before Yap1 redox domain, distinguish Phlebioid clad (Phanerochaetes) of Polyporales from Antrodia clade and all other Basidiomycota tested. In addition to the two domains, high level of identity is found between these Basidiomycota sequences before and after B_ZIP1 domain which confirms the homology between these sequences. A very low level of identity has been found in the sequences localized between the two domains when Ascomycota and Basidiomycota putative homologs are compared and it was not possible to align these intermediate sequences except for region PAP1 chain A (cf. Fig. S3). The position of the beginning of the sequence used for A. bisporus population study is indicated but the end of this sequence is inside the 3’UTR of the gene. A.bi_028, A. bisporus EKV52013; Cci_bZIPfTF, Coprinopsis cinerea B_ZIP family transcription factor XP_001828489; Ppl_bZIPfTF, Postia placenta Prot ID 93814-mod (annotation modified); Pca_bZIPfTF, Phanerochaete carnosa EKM60492; Pch_ bZIPfTF, Phanerochaete chrysosporium Prot ID 7851-mod; Ade_ bZIPfTF, Auricularia delicata EJD55397; Sla_ bZIPfTF, Serpula lacrymans EGN94090. Putative NLS and B_ZIP1 domain of Basidiomycota and Ascomycota homologs of Yap1 and 028-1. a: alignment overview. The B_ZIP1 domain is well conserved among Ascomycota and Basidiomycota. A bipartite NLS motif has been found experimentally on Yap1 (Sce) (blue in first line). We have tried to deduce a putative NLS position in Abi by homology (alignment) of sequences and using NLS Mapper software. For alignment we have found homologs belonging to Saccharomycotina as yeast (Cja, Cte, Cal, Ecy), Pezizomycotina (Mph, Dse, Aol, Tme, Mac, Bfu, Fps, Cmi, Che, Pma, Cim, Aka), Pucciniomycotina (Pgr), Agaricomycotina (Tremellales – Cne, Tms, Tas, Dacrymycetales – Dac, Sebacinales – Pin, Auriculariales – Ade, Polyporales – Fra, Pca, Corticilales – Pst, Russulales – Shi, Boletales – Csu, Sla, Agaricales – Cci, Abi). Intron positions are indicated in Dse, Tme, Che, Tms, Dac and Abi sequences using the convention proposed by Billette et al. [56]. Partial or total transformation of an intron (green diamond in Dse and Che sequences) to an exon could be at the origin of sequence elongation in Basidiomycota on the N-terminal side, just beside the B_ZIP1 domain. In Aka_ bZIPfTF, the sequence VRMLGYRDVGAESLVSLTNSQ corresponds certainly to an error of annotation: It begins by GTACG and finishes by AG on the gene sequence (BACL01000125: 1174-1236) and is exactly at the same place as the intron found on Dse and Che sequences. NLS Mapper score for bipartite NLS (sequences highlighted in red) were 2.6, 3.9, 9.3, 10.2, 8.5, 5.6 and 4 for Sce N-terminal, Sce C-terminal, Pma, Sla, Cci, Abi N-terminal, Abi C-terminal respectively. Pma, Sla, Cci, Abi N-terminal predicted NLS are located at the same position and have the best scores. The consensus sequence LKRKAS, on the N-terminal side of the putative NLS, is also conserved in all 028-1 protein homologs belonging to Polyporales, Corticiales, Russulales, Boletales and Agaricales analyzed here but Coprinopsis cinerea (PKRKAA). For Tremellales, a putative NLS may exist at another position as the Tms sequence RKGRKSGGGEDGDGKGPKEKEMGKAARRK has a score of 6.6 with NLS mapper. NLS Mapper prediction does not fit exactly experimentally identified NLS in yeast but it covers it. This difference exists also for the yeast Pho4p, another target protein of Pse1: SANKVTKNKSNSSPYLNKRRGKPGPDSATSLFELPDS is the experimental determined NLS and KRRGKPGPDSATSLFELPDSVIPTPKPKPKPK is the best cNLS Mapper prediction (score 4.9) for a bipartite NLS. To design the software, Kosugi et al. [35] selected peptides bound by importin alpha. They screened a library of thioredoxin - green fluorescent protein (Trx-GFP) fused with various NLS peptides using a newly developed universal GFP expression system. The NLS activity was scored on a 10-point scale based on GFP localization phenotypes that can be only nuclear (score 10) to cytoplasmic (score 1). They demonstrated that the consensus basic patterns of the classic NLSs are not always required for activity. By example for bipartite NLS, three peptides in Kosugi's article (figure S6A - peptide 222a KRKAEYAHRIAPPSKR and peptide 124a KRRAPFPDENASPSKK figure S6C - peptide 62y RKRTHDELCGRYPSRG) active in yeast are not conform to consensus sequence defined as (K/R)(K/R)X10-12(K/R)3/5 where (K/R)3/5 represent at least three of either lysine or arginine of five consecutive amino acids and X10-12, any 10-12 amino acids. These three peptides have one more (K/R) residue on the N-terminal region and less than three lysines or arginines among the five consecutive amino acids on the C-terminal region. From this analyze, we can deduce that part or the totality of a putative bipartite NLS is present on the Abi sequence RESNLKRKASFGGDNSSNGPSQKSQHTLSNDKKGATSSSAGSRRKSIGG. The Threonine in grey (Thr-93) is replaced by an Isoleucine in Abi var burnettii JB137-S8 genome (Protein Id 144436). Sce_Yap1, Saccharomyces cerevisiae DAA09892, Cja_bZIPfTF, Cyberlindnera jadinii_BAE48266, Cte_bZIPfTF, Candida tenuis_EGV63639, Cal_bZIPfTF, Candida albicans_XP_721702, Ecy_bZIPfTF, Eremothecium cymbalariae_XP_003645961, Mph_bZIPfTF, Macrophomina phaseolina_EKG11847, Dse_bZIPfTF, Dothistroma septosporum_EME48873, Aol_bZIPfTF, Arthrobotrys oligospora_EGX43328, Tme_bZIPfTF, Tuber melanosporum_XP_002837914, Mac_bZIPfTF, Metarhizium acridum_EFY90531, Bfu_bZIPfTF, Botryotinia fuckeliana-CCD34878, Fps_bZIPfTF, Fusarium pseudograminearum_K3V4Q6_FUSPC, Cmi_bZIPfTF, Cordyceps militaris_G3JUL9_CORMM, Che_bZIPfTF, Cochliobolus heterostrophus_Q5J7N6_COCHE, Pma_bZIPfTF, Penicillium_marneffei_XP_002145732, Cim_bZIPfTF, Coccidioides immitis_XP_001247611, Aka_bZIPfTF, Aspergillus kawachii_GAA85896, Pgr_bZIPfTF, Puccinia graminis f. sp. tritici_XP_003320044, Cne_bZIPfTF, Cryptococcus neoformans_XP_778215, Tms_bZIPfTF, Tremella mesenterica_EIW70727, Tas_bZIPfTF, Trichosporon asahii_K1VEC3, Dac_bZIPfTF, Dacryopinax sp._EJT99040, Pin_bZIPfTF, Piriformospora indica_G4TFT1_PIRID, Ade_bZIPfTF, Auricularia delicata EJD55397, Fra_bZIPfTF, Fibroporia radiculosa_CCL99636, Pca_bZIPfTF, Phanerochaete carnosa EKM60492, Pst_bZIPfTF, Punctularia strigosozonata_EIN13303, Shi_bZIPfTF, Stereum hirsutum_EIM83320, Csu_bZIPfTF, Ceriporiopsis subvermispora_EMD41389, Sla_bZIPfTF, Serula lacrymans_EGN94090, Cci_bZIPfTF, Coprinopsis cinerea_XP_001828489, Abi_bZIPfTF, Agaricus bisporus_EKV52013. b and c: detail of the alignment. On the bZIP sequence the five amino acids (N-74, A-77, Q-78, F-81 and R-82 in Sce_Yap1) known to make contact with DNA in Yap family in yeast are conserved in Abi_bZIPfTF and in all the sequences analyzed here but Cte_bZIPfTF and Cal_bZIPfTF where Phenylalanine (F-81) is replaced by a Tyrosine Y as in other members of yeast Yap family (Yap5 and Yap7). The two S. cereviciae Yap family-specific residues, a glutamine Q-73 and an alanine A-80 are conserved (they are replaced by two Arginine R in Gcn4 the typical yeast bZIP transcription factor). This demonstrates that all these sequences belong to Yap family among the bZIP Transcription Factor family (bZIPfTF). In Yap8p N-74 from Yap1p is replaced by a L and A-77 by a S. In the leucine zipper, the typical positions of the coiled coil (Leucine at position d and hydrophobic residues at position a in a helical wheel diagram) are indicated [17]. Sequence alignment of putative chain A of PAP1 region. The most conserved part of chain A is framed and Cysteine positions in Yap1p are specified. The arrows indicate the two Cysteine positions conserved in many bZIPfTP sequences in the three subphyla of the Ascomycota but absent in Basidiomycota species studied. A.bi_ bZIPfTP, A. bisporus EKV52013 (028-1), Sce_Yap1, Saccharomyces cerevisiae DAA09892, Bad_ bZIPfTP, Bjerkandera adusta_jgi _74104 (Protein ID in JGI portal http://genome.jgi-psf.org/programs/fungi/index.jsf), Dsq_ bZIPfTP, Dichomitus squalens_EJF58795, Sco_ bZIPfTP, Schizophyllum commune_EFI91622, Slu_ bZIPfTP, Suillus luteus_jgi _809202, Hir_ bZIPfTP, Heterobasidion irregular_jgi _443323, Scp_ bZIPfTP, Saitoella complicata_jgi_15991, Spo_ bZIPfTP, Schizosaccharomyces pombe_NP_593662, Kla_ bZIPfTP, Kluyveromyces lactis_XP_451077, Yli_ bZIPfTP, Yarrowia lipolytica_XP_504945, Cpo_ bZIPfTP, Coccidioides posadasii_XP_003065816, Agy_ bZIPfTP, Arthroderma gypseum_XP_003169689, Pfi_ bZIPfTP, Pseudocercospora fijiensis_EME88330, Eni_ bZIPfTP, Emericella nidulans_Q5AW17, Afu_ bZIPfTP, Aspergillus fumigatus_Q4WMH0. Other sequence and species names are explained in Supplementary Figs. S1 (Ppl and Pch) and S2. Sequence alignment of the Yap1 redox domain, corresponding to the chain B of PAP1 region. In Yap1p, under oxidizing conditions, the formation of two disulfide bonds (C303-C598 and C310-C629) masks the NES motif [32]. The exportin Crm1 cannot bind Yap1p which is retained in the nucleus and activates the target genes. The Yap1 redox domain and the NES motif are well conserved among Ascomycota for Pezizomycotina (Tme, Pma, Bfu) and among Basidiomycota, better for Agaricomycotina than for Pucciniomycotina (Mla, Cqu) bZIPfTF studied. A Cysteine homologous to Sce_C-629 is present in A. bisporus (C-583), but the formation of a disulfide bond homologous to Yap1P C310-C629 cannot happen as C310 has no homolog in A. bisporus 028-1 (Supplementary Fig. S3). For disulfide bond C303-C598, the region surrounding C-598 on the C-terminal side of Yap1 redox domain is not homologous to the corresponding region of 028-1 and none of these two Cysteines have a homolog in A. bisporus. Another Cysteine (vertical arrow), 17 amino acids forward the Cysteine of Ascomycota, could play in Agaricomycotina the role of C-598 from Sce, with a Cysteine in chain A, 8 positions forward Sce-C303 (Supplementary Fig. S3). Experimental demonstration is needed. Position of Cysteine C-620, involved in thiol reactive agents sensing in Sce, is conserved in 028-1 (C-574). NES motif can be predicted in A. bisporus 028-1 as hydrophobic residues and an acidic residue in between are in conserved positions as in viral and metazoan NES. These positions are conserved in 028-1 basidiomycota homologs studied. Yap1 redox domain is very similar between Yap1p and Yap2p. But a Cysteine Yap2p-C391 (horizontal arrow) involved in Cadmium sensing [33] is absent in Yap1p and in all bZIPfTF Yap1 homologs studied. Mla_ bZIPfTF, Melampsora laricis-populina_jgi_92181, Cqu_ bZIPfTF, Cronartium quercuum fusiforme_jgi_717477. Other sequence and species names are explained in Supplementary Fig. S1, S2 and S4.