1 Introduction
Often highlighted, relationships between polyploidy, morphology, mating systems, fertility and ecological adaptations are still under discussion [1–4]. Whole-genome duplication events may provide high levels of heterozygosity in doubling allele number [5], maintaining genetic diversity at intra-genomic scale [6] and allowing subfunctionalization of duplicate genes [7]. Thus, it has been hypothesized that polyploidy could induce new genomic entities possessing higher adaptability and phenotypic plasticity than their progenitors [8]. For these “super-genotypes”, the maintenance of sexual reproduction could represent a risk due to outbreeding. In addition, the frequent formation of multivalents could cause strong meiotic irregularities leading to non-viable gametes, which dramatically reduces reproductive success [9]. Thus, clonal species may possess a higher competitive ability to persist in changing environments and marginal habitats without investing energy in sexual reproduction [6].
In Poaceae, the mating system of annual crops is well known, because they have been selected, crossed or even polyploidized (e.g., wheat, oat, rye) to increase seed yields since the Neolithic. In contrast, long-lived and tall reedy grasses are also polyploid, but they mainly show low fertility, auto-incompatibility, anemophily [10] and even rare flowering events (Bamboos). The subfamily Arundinoideae Dum. provides a suitable case to assess the impact of polyploidy on reproduction. After removal of the Danthonioid group, it currently remains the smallest grass subfamily, mainly subtropical, with 15 genera and ca. 44 species [11]. All taxa counted are polyploid and the literature suggests x = 6, 9, 12 as chromosome base numbers for Arundinoideae [11]. Following Conert [12], x = 6 seems the most suitable theoretical base number because:
- (i) it characterizes the related subfamily Danthonioideae Barker & H.P. Linder (2n = 12, 24, etc.);
- (ii) it is the only one compatible with the various numbers found in Arundinoideae (2n = 36, 42, 48, 54, 72, 90, 108);
- (iii) it avoids the conflictual consideration of 2n = 36 as a 3x in Phragmites and as a 4x in Molinia, two sister genera.
The genus Phragmites (five species) has been extensively studied [13], particularly P. australis (Cav.) Trin., one of the most widespread taxa. For Lambertini et al. [14], European populations of this common reed “could be considered as member of a single metapopulation” that form monoclonal stands up to 86 ha. Moreover, this polyploid complex [15,16] has everywhere a low mean seed set (< 10%: [17,18]). Eurasian Arundo spp. look like Phragmites spp., resulting from morphological convergences of rhizomatous taxa living in similar wetland habitats. Nevertheless, Arundo spp. are above all ruderal plants growing in waste-lands. Owing to their vast range, dynamism and high biomass, P. australis and A. donax are currently selected for biofuel production [19,20]. Arundo taxa are characterized by tall panicles (up to 60 cm) bearing thousands of protandrous florets, long-time autumnal flowering, pollen and seed dispersion by wind. According to recent phylogenetic studies [21–23], this genus has at least five species across subtropical Eurasia: one endemic to Taiwan, A. formosana Hackel, and four living in the Mediterranean area: the Eurasian A. donax L., the circum-Mediterranean A. micrantha Lam., the Italian-Balkan A. plinii, and the narrow French-Ligurian endemic A. donaciformis (Loisel.) Hardion et al.
Previous morphological and phylogenetic studies have evidenced a failure of seed set in most of these taxa, low intraspecific genetic diversity and two high ploidy levels, 12x and 18x [21–23]. One non-fructiferous genotype of the common A. donax L. (the giant reed) has invaded almost all the world's warm regions [23,24]. For this 18x-clone, severe alterations of gametogenesis prevent its sexual reproduction [25–27], while seed set occurs in Asia [28,29] where A. donax possesses a lower ploidy level 12x [30–32].
The present study assesses the potential impact of polyploidy on the reproductive biology of Mediterranean Arundo. In particular, we test the hypothesis that polyploidy events induce reproductive failures of gametogenesis and seed set. For this purpose, we compare ploidy levels estimated by chromosome counts and DNA content:
- (i) with microscopic observations of male gametogenesis steps, looking for early failures of sexual reproduction;
- (ii) with seed sets in and ex situ, in genotype-isolated and admixture conditions, testing auto-incompatibility phenomena;
- (iii) with the number and the activity of rDNA sites, potentially evidencing restructuration of highly polyploid genome.
2 Materials and methods
2.1 Plant materials
This study concerns the four species of the genus Arundo occurring in the Mediterranean basin. Caryological and morphological analysis was carried out on the basis of an extensive sampling of 91 localities: 16 for A. donax, 27 for A. micrantha, 40 for A. plinii, and 8 for the endemic A. donaciformis (See details in Appendix A). For each locality, five samples (stem with panicle) were deposited in the herbarium of Aix-Marseille University (MARS).
Rhizomes collected in situ were rooted in water to provide clean young material; all accessions were treated under standard conditions. After a cold pre-treatment of 4 °C for one day, root tips were fixed with an ethanol–acetic acid solution (4:1, v/v), kept at room temperature for two weeks, and then stored at –18 °C until used. Similarly, for the gametogenesis study, during the flowering periods (2011 and 2013 autumns), young and mature spikelets were fixed in situ, and/or sampled weekly ex situ. All materials were stained in 45% aceto-carmine-ferriacetate, boiled for 3 min, and then squashed between slide and cover-slide. Two samples per locality were studied and, for each one, the three best mitotic metaphases were identified by light microscopy (15 × 100, Leitz Dialux 20, equipped with a Canon EOS 550D camera). Chromosomes were counted on photographs and drawn using a camera lucida. To complete this study, flow cytometry analysis was performed using internal standards (Petunia hybrida PxPC6, 2C = 2.85 pg) to determine 2C DNA content (Partec CyFlow 532-nm laser cytometer) following the technique described by Fridlender et al. [33].
2.2 Fluorescent in situ hybridization (FISH) of rDNA loci and AgNOR staining
Fixed material was washed in citrate buffer (10 mM, pH 4.6), and macerated for 1 h at 37.8 °C in a solution of 2% (v/v) cellulase (Calbiochem) in citrate buffer, and 20% pectinase in 40% glycerol in citrate buffer. Slides were stained with 4% Giemsa solution diluted with Sörensen phosphate buffer (0.2 M; pH 6.9). The 45S and 5S rDNA multigene families were localised using the pTa71 [34] and pTa794 [35] clones, respectively. The pTa71 and pTa794 probes were labelled with digoxigenin-11dUTP and biotin-16-dUTP, respectively, through a nick translation procedure (Roche, Germany). We followed the in situ hybridization protocols of Rosato et al. [36].
Digoxigenin-labelled probe detection was performed with anti-digoxigenin antibodies conjugated to fluorescein isothiocyanate (Roche). Biotin-labelled probe detection was performed with streptavidin conjugated with Texas Red (Vector Laboratories, Peterborough, UK). Finally, DAPI counterstaining was carried out on slides mounted in Vectashield (Vector Laboratories). Fluorescent signals were detected using an Olympus fluorescence microscope with Camedia C-2000-Z digital camera. The images were optimized for best contrast and brightness on Adobe Photoshop v.7.0. Active NORs and maximum number of nucleoli were identified after silver nitrate staining [37].
2.3 Meiosis and pollen study
From spikelet fixations, meiosis and pollen mitosis were observed on stained anthers of young florets, for each taxon. Owing to the pollen uniformity of Poaceae, only the diameter of mature pollen grains (i.e., equatorial axis) was measured (30 grains per sample) to assess the relationships between ploidy level and cell size. In the botanical garden, at anthesis, pollen was collected from freshly opened florets in the morning and placed in Petri dishes containing a germination medium (1% agar, 15% sucrose, 0.01% boric acid, 0.02% calcium nitrate and 0.02% magnesium sulphate), and incubated at 25 °C for 3 h. Pollen grains were considered germinated when the length of the pollen tube exceeds the grain diameter. Alexander's stain was used to distinguish between abnormal (blue-green coloured) and normal grains (magenta-red stained). The percentage of red stained grains was considered as an indicator of pollen viability [38].
2.4 Self-incompatibility and seed set
Rhizomes collected in situ were transplanted in the Botanical sites of Aix-Marseille University according to two conditions:
- (i) isolated individuals cultivated in greenhouses of Saint-Charles Faculty as control without outbreeding possibilities;
- (ii) admixture of the four species and cytotypes planted in the Botanical Garden of Saint-Jerome Faculty, randomly planted 0.5 m apart from one another.
As reported by Ishii and Kadono [18] for Phragmites, we did not perform emasculation or manual crossing, due to the small size and induration of florets. Seed set rates were estimated for each taxon, on 20 florets per panicle and five panicles per locality.
3 Results
3.1 Chromosome numbers
The synthesis of our counts in the Mediterranean basin exhibits an unexpected chromosomal variability in Arundo (Table 1). This genus is characterized by two ploidy levels and aneuploidy events, except for the French-Ligurian endemic A. donaciformis (2n = 18x = 108). In the other species, we found: 2n = 18x = 108–110 in A. donax, 2n = 12x = 70–72 in A. micrantha, and five cytotypes (2n = 72, 74, 76, 108, and 114) in A. plinii. Whatever the base chromosome number chosen, the multiple 2n = 72 and 108 can be designated as a euploid series, and the others as derived aneuploid numbers. Moreover, only A. plinii exhibited a clear geographical structuring of aneuploid cytotypes (Fig. 1): hyper-aneuploids in central and northern Italy (2n = 76 and 114), euploid numbers in Balkans (2n = 72 and 108), and the strongest diversity in Sicily, with all five cytotypes counted for this species.
Cytogenetic data in Mediterranean Arundo taxa, including ploidy level (2n), DNA content (2C); numbers of 5S and 45S rDNA sites, and maximum number of Ag-staining nucleoli (NORs) observed.
| Taxon | 2n | 2C value (pg) | No. of 5S sites | No. of 45S sites | No. of nucleolus |
| A. micrantha | 12x | 3.2 ± 0.1 | 4 | 4 | 4 |
| A. plinii | 12x | 3.4 ± 0.2 | 4 | 4 | 4 |
| A. plinii | 18x | 4.8 ± 0.2 | 6 | 6 | 5–6 |
| A. donaciformis | 18x | 4.7 ± 0.1 | 5 | 5 | 3 |
| A. donax | 18x | 4.9 ± 0.1 | 6 | 6 | 6 |
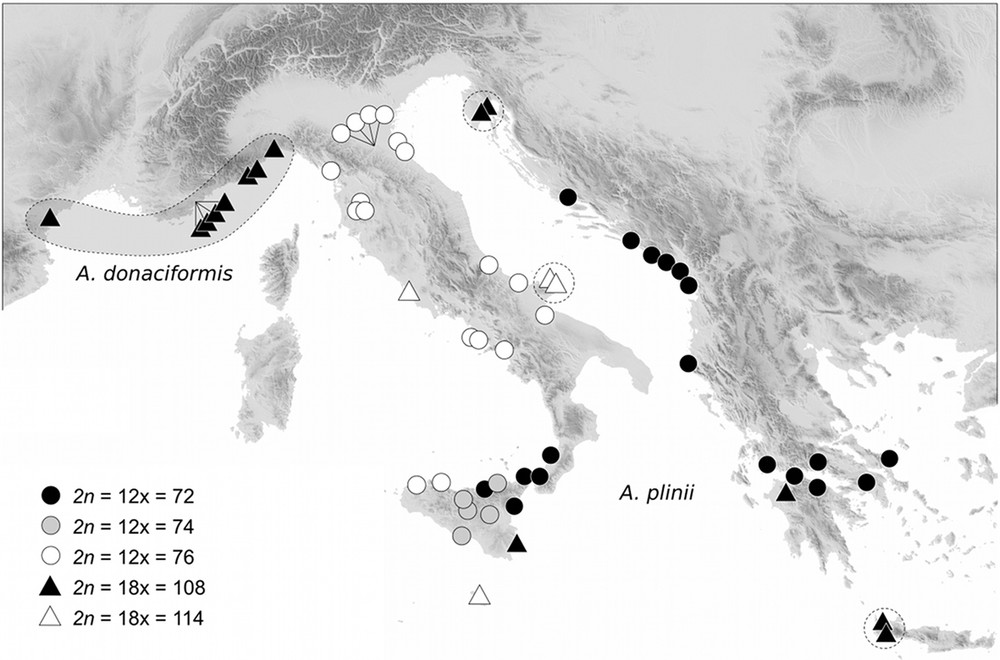
Geographical distribution of the different cytotypes of Arundo donaciformis and A. plinii. Dashed lines group same genotypic clone identified from Hardion et al. [22].
3.2 Polyploidy levels and rDNA sites
The number of rDNA sites (or signals) distinguishes the ploidy levels of Arundo (Table 1). The two 12x-cytotypes, A. plinii (Fig. 2A) and A. micrantha (Fig. 2C), show four 5S and four 45S rDNA sites, whereas six 5S and six 45S rDNA sites are present in the two 18x cytotypes of A. donax (Fig. 2B) and A. plinii. Only the third high polyploid A. donaciformis possesses five 5S and five 45S rDNA (Fig. 2D). AgNOR staining suggests that all 45S rDNA sites are active in both Arundo cytotypes because up to four (Fig. 2G) and six nucleoli (Fig. 2E, F) were observed in the 12x and 18x cytotypes, respectively. As previously, only A. donaciformis 18x is distinguished from all others exhibiting only three nucleoli (Fig. 2H).
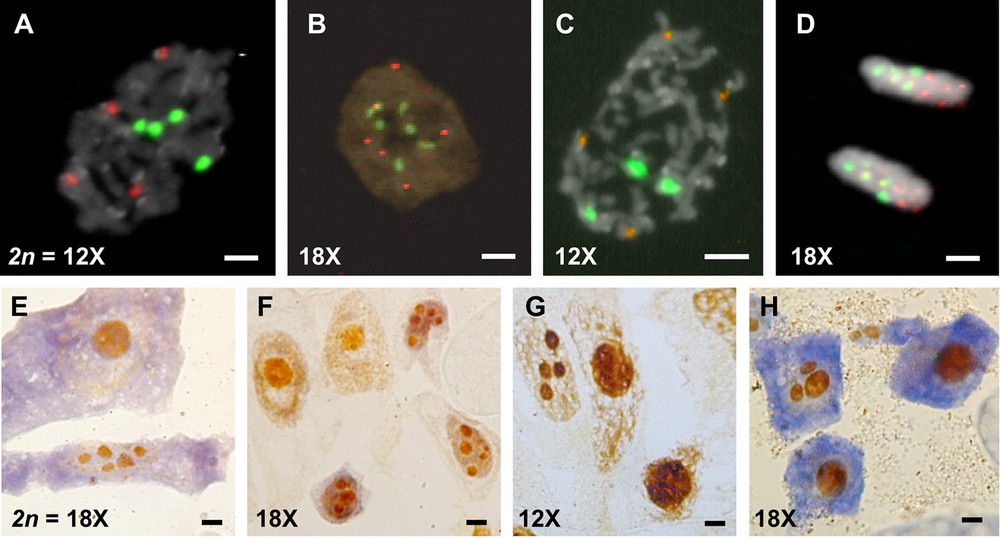
(Colour online.) Fluorescent in situ hybridization (FISH) of 5S rDNA (red) and 45S rDNA (green) probes on cells counterstained with DAPI (A, B, C, D), and number of nucleoli using Ag-staining on somatic cells (E, F, G, H). A. Prophase of A. plinii 12x: four 5S and four 45S rDNA sites. B. Interphase of A. donax 18x: six 5S and six 45S sites; C. Metaphase of A. micrantha 12x: four 5S and four 45S sites. D. Telophase of A. donaciformis 18x: five 5S and five 45S sites. E. A. plinii 18x: up to six nucleoli. F. A. donax 18x: up to six nucleoli. G. A. plinii 12x: up to four nucleoli. H. A. donaciformis 18x: up to three nucleoli. Scales: white bars = 2 μm, black bars = 1 μm.
3.3 Meiosis and pollen study
Microgametogenesis would appear to be regular in most Arundo taxa (Fig. 3A and B), therefore the observation of cytomixis events (i.e. chromosome exchange between two mother cells) in A. micrantha (12x; Fig. 3C) reveals the presence of severe disturbances. Pollen mitoses generally appear regular (Fig. 3E, F, G), but sometimes in the 18x cytotypes deviant haploid numbers ranging from 50 to 62 could be observed. Large size grains (macro-pollen) were found for each cytotype, assumed to correspond to unreduced gametes (2n gametes; Fig. 3J).
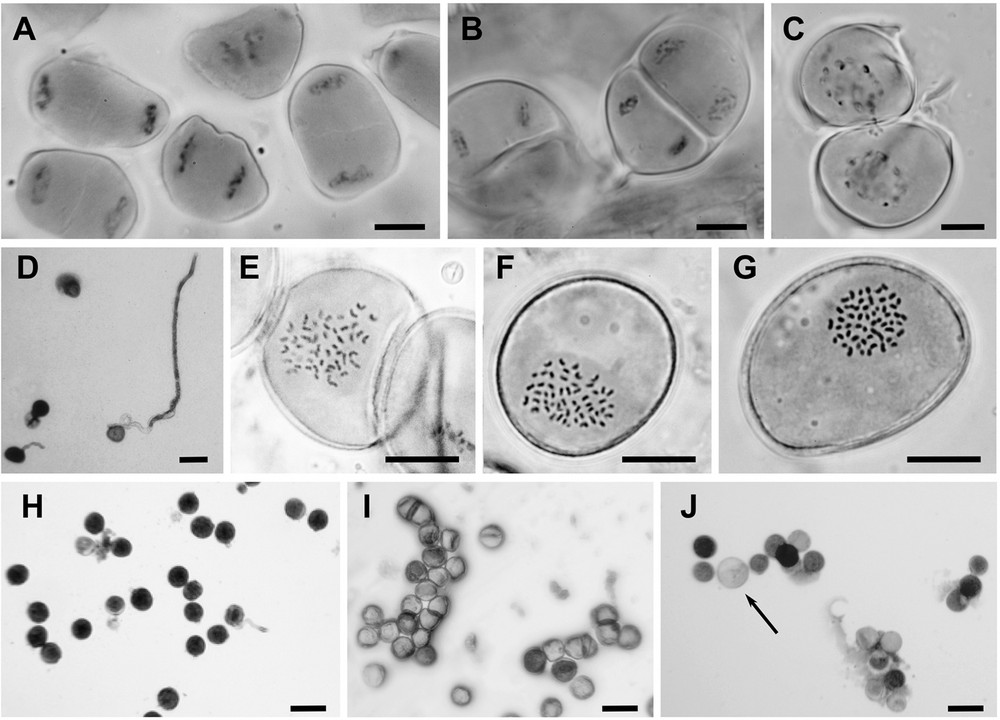
Observations of meiosis (A, B, C), mitosis (E, F, G), viability (H, I, J) and germination (D) of pollen. A. Telophase I of A. plinii 12x. B. Anaphase II of A. plinii 12x. C. Cytomixis of A. micrantha 12x: chromosome exchange between two mother cells. D. Pollen germination of A. plinii 18x. E. Normal pollen and pollen mitosis of A. donaciformis 18x: n = 54. F. Pollen mitosis of A. plinii 18x: n = 56. G. Pollen mitosis of A. donax 18x: n = 54. H. Normal pollen of A. plinii 12x (homogeneous size and staining). I. Collapsed and uncoloured pollen grain of A. donax 18x. J. Abnormal pollen of A. micrantha 12x (heterogeneous in size and colour; arrow showing the diplogamete). Scales: A to H, black bars = 10 μm; H, I and J, black bars = 10 μm.
Whereas pollen stainability values reached 80.8, 84.6% and 86.8% in A. donaciformis 18x and the two cytotypes 12x and 18x of A. plinii, respectively, their pollen germination rates dropped to 10.6%, 7.8%, and 8.0%, respectively (Fig. 3D). In these taxa, pollen stainability was not closely related to germination rate (Table 2). In contrast, both A. micrantha 12x (Fig. 3 J) and A. donax 18x (Fig. 3I) showed similar very low pollen stainability (3.7% and 6.2%, respectively), numerous collapsed grains and no pollen germination. In addition, at the anthesis, these two species show a high frequency of sclerified and empty anthers (84.1 and 91.3%, respectively).
Cytogenetic data including ploidy levels (2n) and haploid numbers from meiosis or pollen mitosis (n); pollen viability and in vitro germination percentages; seed set percentages in situ and ex situ (in isolated or admixed conditions) in Mediterranean Arundo taxa.
| Taxon | 2n | n | Pollen viability (%) | Pollen germination (%) | Seed set (%) | ||
| In situ | Ex situ isolated | Ex situ admixed | |||||
| A. micrantha | 12x | 36 | 3.7 | 0 | 0 | 0 | 0 |
| A. plinii | 12x | – | 84.6 | 7.8 | 9.2 | 0 | 7.3 |
| A. plinii | 18x | 56 | 86.8 | 8.0 | 0 | 0 | 5.2 |
| A. donaciformis | 18x | (50) 54 (62) | 80.8 | 10.6 | 0 | 0 | 6.4 |
| A. donax | 18x | 54–55 | 6.2 | 0 | 0 | 0 | 0 |
3.4 Seed production
In situ, seed set was strictly restricted to the A. plinii 12x, with an average of 9.2%, and was never found in A. micrantha 12x, or within all 18x cytotypes: A. donax, A. donaciformis and A. plinii (Table 2). However, ex situ genotypic admixture in our botanical garden induced the formation of some fructiferous florets (5 to 7%) in the sister species A. donaciformis 18x and A. plinii 12x and 18x, whereas A. micrantha 12x and A. donax 18x remained non-fructiferous. In addition, all plant controls grown in isolation never produced seed.
4 Discussion
4.1 Cytogenetic study
Chromosome counts exhibited two ploidy levels in Arundo taxa, 12 and 18x, with aneuploid variants around each euploid number. Measurements of DNA content support these two ploidy levels, but fail to detect aneuploid events as it was found in other taxa with small genome sizes [39]. Our results confirm previous reports in the Mediterranean area for A donax: 2n = 110 [40,41], 112 [42], and 2n = 72 in A. micrantha from Portugal [43] (under the name A. plinii s.l.) and in A. plinii from southern Italy and Sicily [40]. Generally aneuploids are randomly distributed, often due to difficult counts of these numerous and small chromosomes (0.9–1.9 μm). On the other hand, chromosomal variations show a clear geographical structuring only in the Italian-Balkan A. plinii (Fig. 1). The more ancestral cytotype remains 2n = 72, from which were derived aneuploids (2n = 74–76) in Italy and high polyploid (2n = 108–114) numbers. This geographical pattern did not yet corresponds to previous taxa recently fallen in synonymy of A. plinii, as A. collina Ten. or A. hellenica Dainin et al. [21,44,45]. As in many other groups [46], all 18x cytotypes Arundo probably resulted from an intraspecific fusion of reduced and unreduced gametes, and not from hybridization as suggested [40,41] for the monophyletic A. donax [23]. The rate of diplogametes generally increases with thermal shocks [47], and thus we found A. plinii 18x mainly on the northern and southern edges of the species’ distribution range. According to previous genetic studies, the origin of A. donaciformis 18x was included in the phylogeny of A. plinii [22]. This euploid cytotype (2n = 108) cannot be derived from the nearest northern Italian aneuploids 12x (Fig. 1), but rather from a euploid lineage (2n = 72) conserved in southern Italy and Sicily.
If the most frequent numbers of 45S rDNA sites were two and four in diploid flowering plants [48], in the Poaceae this number is generally two [49,50]. The presence of four functional 45S rDNA sites in 12x Arundo shows a drastic turnover by their physical elimination from ancestral genomes. Except for A. donaciformis, the mathematical increase of 45S and 5S rDNA sites evidenced from 12x to 18x Arundo cytotypes (four and six sites respectively) suggests that this rDNA pattern is evolution-conserved. According to Roa and Guerra [48], these 18x Arundo cytotypes may have a more recent origin than A. donaciformis 18x that shows a trend to reduce the number of rDNA sites (five) and nucleoli (three).
4.2 Gametogenesis and pollen
The cytological impact of polyploidy (e.g. multivalent formation and segregation irregularities) is often designated as a probable cause of reproductive failure in A. donax [26]. If meiosis and pollen mitosis observations seem to show a correct pattern of behaviour, in fact cytomixis events found in A. donax 18x [40] and A. micrantha 12x (in this study) explain male anomalies. This phenomenon is responsible for asymmetric segregation, chromosome laggards and diplogametes that produce aneuploid and polyploid numbers [51,52]. In addition, these two species with distinct ploidy levels showed a very low rate of viable pollen and no germination. Whatever their ploidy levels, only taxa capable of pollen germination can produce seed set ex situ: 12x and 18x cytotypes of A. plinii, and A. donaciformis. These results suggest that microgametogenesis steps may explain the differential reproductive success of Arundo taxa, and discredit the hypothesis of the predominant role of polyploidy in the failure of meiosis and pollen germination.
These features must be related to previous studies on megagametogenesis, with distinct results according to species. A. plinii showed embryo sacs developing in mature ovules, then seeds [26]. In contrast, early megagametophyte degeneration has been reported in Phragmites [53] and A. donax [26,27]. On the basis of 36 666 florets, Johnson et al. [25] found only 43 ovules apparently matured, but only five displaying dehydrogenase activity. In fact, the low seed set found in rhizomatous Poaceae may be mainly due to callose deposition around embryo sacs, producing abnormal pistils with a low capacity to capture pollen grains [54].
4.3 Breeding systems and polyploidy in Arundo
The absence of seed set in isolated plant controls shows that Arundo spp. are not autogamic or agamospermic, conversely to the rule for many polyploids [10]. Knowing that A. donax, A. micrantha and A. donaciformis are monoclonal in the Mediterranean area [21–23], the more probable reason to explain seed set absence in situ is auto-incompatibility processes in the genus, such as evidenced in Phragmites [18]. This feature also explains why only A. plinii 12x produces seeds in situ, whereas related taxa A. plinii 18x and A. donaciformis 18x were only able to produce seeds under genotypic admixture conditions. Thus, polyploidy is not entirely responsible for seed set absence due to self-incompatibility phenomena, geographical distances between populations become preponderant (Fig. 1). This mechanism is reinforced by cytogenetic barriers between 12x and 18x, geographical isolation of all 18x cytotypes and the higher genetic diversity of 12x A. plinii [22]. In this complex, we did not find aneuploidy impact. On the other hand, the low seed set (< 10%) of Arundo is also observed in many other long-lived or rhizomatous plants, such as P. australis [16,18]. Within these taxa, the low seed set generally linked to ovule abortion is compensated by thousands of florets per panicle, allowing resource saving and sufficient regeneration [55].
Finally, there is a paradox concerning the ranges of distribution and reproduction biology of Arundo species: native and potentially fertile taxa, such as A. plinii and A. donaciformis possess smaller ranges of distribution than robust widespread and non-fructiferous clones, such as A. micrantha and A. donax. As already evidenced for A. donax [23], the Mediterranean-wide distribution and the genetic uniformity of these two taxa could be explained by their ancient anthropogenic dispersion due to their countless uses during Antiquity (e.g. arrow and vineyard reeds respectively), as listed by Pliny the Elder. However, the failures of their gametogenesis remain poorly understood. In contrast to speculation in the literature, polyploidy events do not seem to influence their reproductive problems. Related to human impact, these failures could be linked to drastic founder effects and genetic impoverishment of degenerated clones cultivated for millennia, forming a clonal patch without reproductive effort. These early failures of sexual reproduction could also be due to the bioclimatic disequilibrium of these transplanted clones in the Mediterranean [10]. In the context of global change, further investigations should test these environmental vs genetic factors regarding the current sterility of these clonal and invasive species.
Acknowledgements
This work is part of an Interdisciplinary Program of Research Urban-Environment (PIRVE) financed by the CNRS, the Ministry of Ecology (MEDAD, France) and the DREAL PACA. The authors are grateful for other financial support provided by the Research Federation ECCOREV and the Société botanique de France.
Appendix A
| Taxon | Specimen | 2n | DNA content (pg) |
| A. donax | Croatia, Pag, Fridlender, MARS00047 | 108–110 | – |
| A. donax | France, Bonifacio, Verlaque, MARS00048 | 108–110 | – |
| A. donax | France, Marseille, Verlaque, MARS00049 | 108–110 | 4.5 |
| A. donax | Greece, Kolimpari, Vila, MARS00050 | 108–110 | 4.7 |
| A. donax | Greece, Kavassilas, Hardion & Vila, MARS00051 | 108–110 | – |
| A. donax | Italy, Volterra, Hardion & Vila, MARS00052 | 108–110 | – |
| A. donax | Italy, Naples, Hardion & Vila, MARS00053 | 108–110 | – |
| A. donax | Italy, Cannitello, Hardion, MARS00054 | 108–110 | – |
| A. donax | Italy, Carloforte, Fridlender, MARS05001 | 110 | – |
| A. donax | Lebanon, Jadra, Abdel Samad, MARS00055 | 108–110 | – |
| A. donax | Malta, Marsaskala, Médail, MARS00056 | 108–110 | – |
| A. donax | Morocco, Rabat, Hardion, MARS00057 | 108–110 | – |
| A. donax | Portugal, Figueira da foz, Engels, MARS00058 | 108–110 | – |
| A. donax | Spain, Bonares, Hardion, MARS00059 | 108–110 | – |
| A. donax | Spain, Amposta, Hardion, MARS00060 | 108–110 | 4.8 |
| A. donax | Tunisia, Sidi bou said, Hardion, MARS00061 | 108–110 | 4.7 |
| A. donaciformis | France, Fréjus, Hardion, MARS00062 | 108 | 4.7 |
| A. donaciformis | France, Puget, Hardion, MARS00064 | 108 | 4.7 |
| A. donaciformis | France, Les Arcs, Hardion, MARS00065 | 108 | 4.7 |
| A. donaciformis | France, Saint Raphaël, Hardion, MARS00063 | 108 | 4.7 |
| A. donaciformis | France, Lespignan, Hardion, MARS00066 | 108 | 4.6 |
| A. donaciformis | Italy, Cervo, Hardion, MARS00067 | 108 | – |
| A. donaciformis | Italy, Andorra, Hardion, MARS00068 | 108 | – |
| A. donaciformis | Italy, Finale Ligure, Hardion, MARS00069 | 108 | – |
| A. micrantha | Algeria, Tipasa, Baumel, MARS00070 | 70–72 | 3.3 |
| A. micrantha | Algeria, Tizi Ouzou, Ait-Saïd, MARS00071 | 72 | – |
| A. micrantha | Algeria, Tlemcen, Youssef, MARS00072 | 72 | – |
| A. micrantha | Algeria, Sidi Aich, Vela, MARS00073 | 72 | – |
| A. micrantha | France, Golfe Juan, Hardion, MARS00075 | 72 | – |
| A. micrantha | France, Ste Lucie, Argagnon, Michaud & Molina, MARS00076 | 72 | – |
| A. micrantha | Greece, Kissamos, Vila, MARS00077 | 70–72 | 3.1 |
| A. micrantha | Greece, Mires, Vila, MARS00078 | 72 | – |
| A. micrantha | Greece, Mirtos, Vila, MARS00079 | 72 | – |
| A. micrantha | Greece, Vassiliko, Hardion & Vila, MARS00080 | 72 | – |
| A. micrantha | Greece, Messolonghi, Hardion & Vila, MARS00081 | 72 | – |
| A. micrantha | Greece, Itea, Hardion & Vila, MARS00082 | 72 | – |
| A. micrantha | Italy, Trieste, Hardion, MARS00083 | 72 | – |
| A. micrantha | Italy, Las Plassas, Fridlender, MARS00084 | 72 | 3.2 |
| A. micrantha | Lebanon, Nahr el Kalb, Abdel Samad, MARS00085 | 72 | – |
| A. micrantha | Morocco, Tanger, Hardion, MARS00086 | 72 | – |
| A. micrantha | Morocco, Rabat, Hardion, MARS00087 | 72 | – |
| A. micrantha | Israel, Nahal Sorek, Danin, MARS0007 | 72 | – |
| A. micrantha | Portugal, Figueira da Foz, Engels, MARS00089 | 72 | – |
| A. micrantha | Spain, Amposta, Hardion, MARS00090 | 72 | – |
| A. micrantha | Spain, Deltebre, Hardion, MARS00091 | 72 | – |
| A. micrantha | Spain, Jaen, Hardion, MARS00092 | 72 | – |
| A. micrantha | Spain, Cordoba, Hardion, MARS00093 | 72 | – |
| A. micrantha | Spain, Algeciras, Fridlender, MARS03840 | 72 | 3.2 |
| A. micrantha | Spain, Huelva, Hardion & Sánchez-Gullón, MARS00094 | 72 | – |
| A. micrantha | Spain, Huelva, Hardion & Sánchez-Gullón, MARS00095 | 72 | – |
| A. micrantha | Spain, Bonares, Hardion, MARS00096 | 72 | – |
| A. plinii s.s. | Croatia, Brsec, Hardion, MARS00099 | 108 | – |
| A. plinii s.s. | Greece, Sirili, Vila, MARS00100 | 108 | 4.6 |
| A. plinii s.s. | Greece, Lauzakies, Vila, MARS00101 | 108 | – |
| A. plinii s.s. | Greece, Paralia Kimis, Hardion & Vila, MARS00102 | 72 | – |
| A. plinii s.s. | Greece, Sicyone, Hardion & Vila, MARS00103 | 72 | – |
| A. plinii s.s. | Greece, Kavassilas, Hardion & Vila, MARS00104 | 108 | – |
| A. plinii s.s. | Greece, Patras, Hardion & Vila, MARS00105 | 72 | 3.4 |
| A. plinii s.s. | Greece, Messolonghi, Hardion & Vila, MARS00106 | 72 | – |
| A. plinii s.s. | Greece, Levadia, Hardion & Vila, MARS00107 | 72 | – |
| A. plinii s.s. | Greece, Agios Stephanos, Hardion & Vila, MARS00108 | 72 | – |
| A. plinii s.s. | Italy, Napoli, Hardion & Vila, MARS00109 | 76 | 3.3 |
| A. plinii s.s. | Italy, Pozzuoli, Hardion & Vila, MARS00110 | 76 | – |
| A. plinii s.s. | Italy, Candela, Hardion & Vila, MARS00111 | 76 | – |
| A. plinii s.s. | Italy, Rodi Garganico, Hardion & Vila, MARS00112 | 114 | 4.6 |
| A. plinii s.s. | Italy, Termoli, Hardion & Vila, MARS00113 | 76 | – |
| A. plinii s.s. | Italy, Lanciano, Hardion & Vila, MARS00114 | 76 | – |
| A. plinii s.s. | Italy, Ostia, Hardion & Vila, MARS00115 | 114 | – |
| A. plinii s.s. | Italy, Pisa, Hardion & Vila, MARS00116 | 76 | – |
| A. plinii s.s. | Italy, Montegabbro, Hardion & Vila, MARS00117 | 76 | – |
| A. plinii s.s. | Italy, San Giminiano, Hardion & Vila, MARS00118 | 76 | – |
| A. plinii s.s. | Italy, Volterra, Hardion & Vila, MARS00119 | 76 | 3.1 |
| A. plinii s.s. | Italy, Bologna, Hardion & Vila, MARS00120 | 76 | 3.3 |
| A. plinii s.s. | Italy, Castel Maggiore, Hardion & Vila, MARS00121 | 76 | 3.2 |
| A. plinii s.s. | Italy, Sasso Marconi, Hardion & Vila, MARS00122 | 76 | 3.2 |
| A. plinii s.s. | Italy, Pianoro, Hardion & Vila, MARS00123 | 76 | 3.2 |
| A. plinii s.s. | Italy, Forli, Hardion & Vila, MARS00124 | 76 | – |
| A. plinii s.s. | Italy, Ravenne, Hardion & Vila, MARS00125 | 76 | – |
| A. plinii s.s. | Italy, Santo Stefano, Hardion, MARS00126 | 72 | – |
| A. plinii s.s. | Italy, Oliveto, Hardion, MARS | 74 | – |
| A. plinii s.s. | Italy, Orto Liuzzo, Hardion, MARS00127 | 72 | – |
| A. plinii s.s. | Italy, Cannitello, Hardion, MARS03395 | 72 | – |
| A. plinii s.s. | Italy, Dinami, Hardion, MARS00128 | 72 | 3.6 |
| A. plinii s.s. | Italy, Siracusa, Hardion, MARS00129 | 108 | 4.8 |
| A. plinii s.s. | Italy, Enna, Hardion, MARS00130 | 74 | – |
| A. plinii s.s. | Italy, Agrigente, Hardion, MARS00131 | 74 | 3.5 |
| A. plinii s.s. | Italy, Scilliato, Hardion, MARS00132 | 74 | – |
| A. plinii s.s. | Italy, Castellamare, Hardion, MARS00133 | 76 | 3.6 |
| A. plinii s.s. | Malta, Girgenti, Gambin, MARS00134 | 114 | 5.0 |
| A. plinii s.s. | Italy, Salerno, Fridlender, MARS03806 | 76 | 3.3 |
| A. plinii s.s. | Italy, Petralia, Fridlender, MARS05011 | 76 | 3.4 |


