1 Introduction
The genus Sorghum L., a member of the family Poaceae, includes many species, among which Sorghum bicolor (L.) Moench; 2n = 2 = 20, which is an important world crop, used as food and fodder and in sorghum syrup or sorghum molasses, while other species of Sorghum are used as fodder, alcoholic beverages, and biofuels [1–3]. Recently, sorghum has received significant attention because of its newer use as a biofuel feedstock [4,5]. Sorghum plants are mostly cultivated in countries with warmer climates. Most varieties are drought- and heat-tolerant C4 plants and are especially important in arid regions, where the grain is one of the staples for poor and rural people [6–8]. Sorghum can be grown in environments where other crops cannot be able to be grown successfully [9]. Increased demand for limited fresh water supplies, increasing use of marginal farmland, and global climatic trends, all suggest that dry land crops such as sorghum will be of growing importance to feed the world's expanding populations [10].
Genetic diversity of sorghum could be influenced by human factors, including markets and government policies related to land ownership and natural factors such as altitude, soil, climate and gene flow with wild sorghum [10–13]. Studies have been done on the genetic diversity of sorghum by using different types of markers such as Random Amplified Polymorphic DNA (RAPDs) [14,15] and microsatellites [16]. A highly significant genetic variation both within and among accessions of sorghum from Zambia and Botswana, respectively, were reported using SSR markers [17,18]. Recently, the relationships and genetic distance among sorghum varieties have been evidenced by using ISSR fingerprinting as a more reliable molecular marker compared to RAPD [19–21].
Assessing the level of genetic diversity among sorghum genotypes as revealed by ISSR analysis has a great relevance for breeding programs [22]. The genetic characterization of landraces makes them an important resource as potential donors of genes for the development of new crop varieties. The potential of ISSR markers to generate genetic information depends on the inter-microsatellite frequency and their distribution in the genome wide scale of the species. The aim of the present study was to produce ISSR markers for the identification of sorghum landraces and determine the genetic relationships among different genotypes growing in Saudi Arabia and Yemen.
2 Materials and methods
2.1 Plant material
Fifteen different accessions of sorghum were collected from different farms in various regions of the Kingdom of Saudi Arabia and Yemen. The codes and areas for the accessions are given in Table 1. Sorghum grains were germinated in the greenhouse and young fresh leaf samples of each samples were packed separately in polyethylene bags, immediately placed in an ice box and transferred to the laboratory for storage at −70 °C until used for DNA extraction and ISSR fingerprinting processing.
List of the 15 sorghum accessions studied and their geographic distribution in Saudi Arabia and Yemen.
| Serial | KACST ID number | Region and/or area | Comments |
| 1. | 01 | Al-Baha, Mekhwat | White grains |
| 2. | 02 | Al-Baha | White grains |
| 3. | 07 | Jazan, Al-Hashr, Mountain | White grains |
| 4 | 08 | Abha | Abo Shoka grains |
| 5. | 113 | Jazan, Al-Hashr, Mountain | Yellow grain |
| 6. | 66 | Jazan, Telab Mountain | White grains |
| 7. | 129 | Abha | Red grains |
| 8. | 215 | Al-Qasim, Al-Badaie | White grains |
| 9. | 216 | Al-Qasim, Unaizah | White grains |
| 10. | 217 | Yemen, Ab | Dark grains |
| 11. | 197 | Yemen, Ab | Red grains |
| 12. | 242 | Yemen, Ab, Jelly | Red grains |
| 13. | 243 | Yemen, Ab, Jelly | Yellow grains |
| 14. | 248 | Yemen, Ab, Jelly | White grains |
| 15. | 249 | Yemen, Ab | Yellow grains |
2.2 DNA Extraction
The leaves were first ground into a fine powder in liquid nitrogen using a pestle and a mortar and DNA was extracted following the steps of Dellaporta et al. [23] with some modifications. Using a fluorometer (Hoefer DNA Quant 200; the quantity and quality of the DNA were determined. Total DNA isolation was also run on a 1% agarose gel to determine consistency and purity. The stock DNA samples were diluted with sterile TE buffer to make a working solution of 10 ng·ml-1 for use in PCR analysis.
2.3 ISSR Fingerprinting
A total of 20 ISSR primers synthesized by ‘IDT, Integrated DNA technologies’ were used for PCR. The primers were dissolved in sterilized distilled water at a concentration of 10 pmol/μl. Amplification reactions were performed in a 25-μl volume using the “Ready-To-Go PCR Beads” kit (GE Healthcare Life Sciences) containing thermostable polymerases—2.5 units of recombinant pure Taq DNA polymerase, dNTPs (200 μM each dNTP and buffer [1.5 mM MgCl2, 50 mM KCl and 10 mM Tris (pH 9)]). In each reaction, 25, ng of DNA samples were used along with 20 pmol/μl of primer. PCR amplification was performed in an Eppendorf Master Cycler Gradient PCR machine. The following PCR program was used:
- 1) one cycle for 5 min at 95 °C;
- 2) 35 cycles at 94 °C for 1 min, 45–55 °C (depending on the Tm of the primers) for 1 min and 72 °C for 2 min;
- 3) one cycle for 10 min at 72 °C, followed by soaking at 4 °C.
The ISSR products were separated by electrophoresis according to their molecular weight on 1.4% (w/w) agarose gels submerged in 1 × TBE buffer and then stained with an ethidium bromide (10 mg·ml−1) solution for 20 min. The DNAs were visualized on a UV trans-illuminator and documented by using the Gel Documentation System of Alpha Innotech. Multi Imaging. To ensure the reproducibility and reliability of the ISSR markers, we repeated the PCR reactions twice with each primer. The primers that showed weak or no patterns were discarded. The length of the amplified ISSR fragments was estimated by running 100 pb Ladder (Bio Rad) in the gel as a standard size marker.
2.4 Data analysis
Since ISSR primers are dominant markers, amplified bands were scored for the presence (1) or the absence (0) based on Nei's genetic distance as implemented in the NTSYS-pc, and the genetic distance among the genotypes was expressed as a distance tree by using the NTSYS-pc software using the unweighted pair-group method with arithmetic averages (UPGMA) and a simple matching coefficient [24]).
3 Results and discussion
Different primers produced different level of polymorphism among the 15 sorghum genotypes; examples of the ISSR fingerprinting profiles are illustrated in Fig. 1. The number of polymorphic bands per primer varied between 6 and 23. A total of 92 alleles were amplified with an average of 13 polymorphic ISSR alleles per primer. ISSR primers numbered 4, 6, and 7 produced a higher percentage of polymorphic fragments (100%); the latter primer produced exceptionally the high number of 23 alleles. All of the seven selected primers produced more than 67% of polymorphic fragments, except the primer coded 1, which showed 16.66% polymorphism; this primer produced the least number of alleles compared to the six other primers (Table 2).
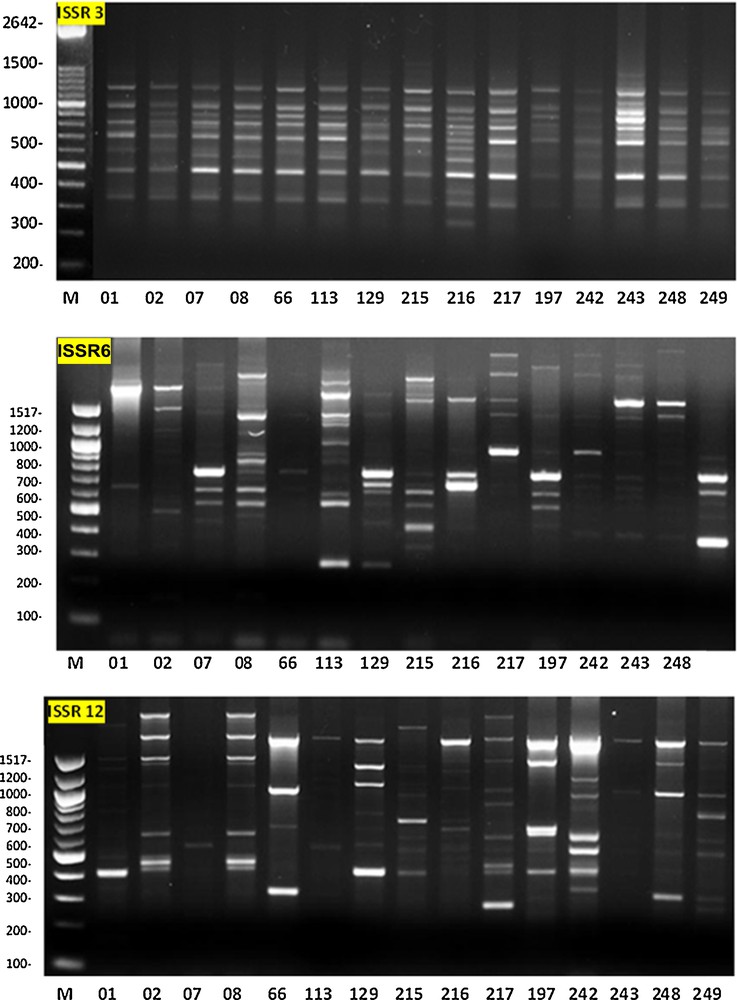
Photographs illustrating the ISSR fingerprinting in 15 genotypes of sorghum by primer 3, primer 6, and primer 12 (see Table 2 for primers sequence).
The number of amplified polymorphic and monomorphic bands band and total percentage of polymorphism as revealed by seven ISSR primers in 15 genotypes of sorghum representing different landraces in Saudi Arabia and Yemen.
| ISSR Primers | Primer sequence | No. of bands | Polymorphic Bands | Monomorphic Bands | Polymorphism % |
| ISSR1 | (AG)8 C | 06 | 1 | 5 | 16.66% |
| ISSR 2 | AG)8 G | 12 | 8 | 4 | 66.67% |
| ISSR 3 | (GA)8 T | 13 | 9 | 4 | 69.23% |
| ISSR 4 | (GA)8 A | 17 | 17 | 0 | 100.0% |
| ISSR 5 | (TC) 8 C | 11 | 10 | 1 | 90.90% |
| ISSR 6 | (TG)8 A | 10 | 10 | 0 | 100.0% |
| ISSR 7 | (CTC)6 | 23 | 23 | 0 | 100.0% |
| Total | 92 | 78 | 14 | 84,78 |
The pairwise genetic distance estimates of the 15 examined sorghum landraces were analyzed and are given in Table 3. The similarity matrix values ranged from 0.436 to 0.818. Maximum similarity was observed between the sorghum genotypes with KACST ID 01 and KACST ID 02 (0.818), both white grain landraces from Al Baha in Saudi Arabia, as shown in Table 1. The second highest similarity was between genotype KACST ID 248 that has red grains and KACST ID 249 that has yellow grains; both landraces from Ab in North Yemen (0.800) and the third highest similarity was between the two landraces KACST ID 129 from Abha and KACST ID 197 from Ab in Yemen; both have red grains. Higher values were also found between the three landraces KACST ID 249, KACST ID 129, and KACST ID 197 (Table 3). The average similarity coefficient among all the 15 individuals was more than 50%.
Similarity matrix expressed as Nei's coefficients among the 15 genotypes of sorghum as revealed by seven ISSR primers (See Tables 1 and 2).
| Code # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1 | 1 | 1.000 | ||||||||||||||
| 2 | 2 | 0.818 | 1.000 | |||||||||||||
| 7 | 3 | 0.763 | 0.654 | 1.000 | ||||||||||||
| 8 | 4 | 0.600 | 0.709 | 0.618 | 1.000 | |||||||||||
| 66 | 5 | 0.672 | 0.636 | 0.763 | 0.527 | 1.000 | ||||||||||
| 113 | 6 | 0.654 | 0.581 | 0.709 | 0.690 | 0.690 | 1.000 | |||||||||
| 129 | 7 | 0.709 | 0.636 | 0.581 | 0.745 | 0.600 | 0.727 | 1.000 | ||||||||
| 197 | 8 | 0.636 | 0.600 | 0.618 | 0.745 | 0.636 | 0.654 | 0.781 | 1.000 | |||||||
| 215 | 9 | 0.709 | 0.636 | 0.618 | 0.600 | 0.636 | 0.509 | 0.709 | 0.636 | 1.000 | ||||||
| 216 | 10 | 0.490 | 0.490 | 0.436 | 0.527 | 0.527 | 0.472 | 0.527 | 0.527 | 0.672 | 1.000 | |||||
| 217 | 11 | 0.563 | 0.563 | 0.581 | 0.709 | 0.563 | 0.618 | 0.745 | 0.745 | 0.709 | 0.672 | 1.000 | ||||
| 242 | 12 | 0.636 | 0.563 | 0.581 | 0.636 | 0.600 | 0.654 | 0.672 | 0.636 | 0.636 | 0.600 | 0.672 | 1.000 | |||
| 243 | 13 | 0.490 | 0.454 | 0.472 | 0.600 | 0.563 | 0.618 | 0.636 | 0.600 | 0.636 | 0.636 | 0.672 | 0.672 | 1.000 | ||
| 248 | 14 | 0.581 | 0.618 | 0.454 | 0.618 | 0.654 | 0.600 | 0.690 | 0.654 | 0.618 | 0.654 | 0.618 | 0.581 | 0.690 | 1.000 | |
| 249 | I5 | 0.563 | 0.527 | 0.581 | 0.709 | 0.672 | 0.690 | 0.781 | 0.781 | 0.636 | 0.527 | 0.709 | 0.600 | 0.745 | 0.800 | 1 |
The cluster analysis of ISSR polymorphism by the UPGMA method showed two main groups, A and B (Fig. 2). Group A was comprised of five landraces with white grains from Jazan and Abha, with a similarity coefficient range from 0.527 to 0.818. Group B was comprised of 10 landraces, with a similarity rate of 0.490 to 0.709. Two genotypes from Al-Qassim in the Middle of Saudi Arabia (KACST ID 215 and KACST ID 216) were clearly delimited from the remaining eight samples. The eight genotypes were divided into two clusters; one was comprised of landraces with dark grains from Abha in Saudi Arabia and Ab in Yemen with a similarity range from 0.563 to 0.781. These samples have KACST IDs 8, 129, 197 and 217). The other four landraces were differentiated into three white colored grains genotypes (KACST IDs 243, 248 and 249) as one subcluster and one dark-colored grain genotype (KACST ID 242); all from North Yemen with a similarity range of 0.454–0.800. The current results encourage further collection and authentication of sorghum landraces in the gene bank of Saudi Arabia.
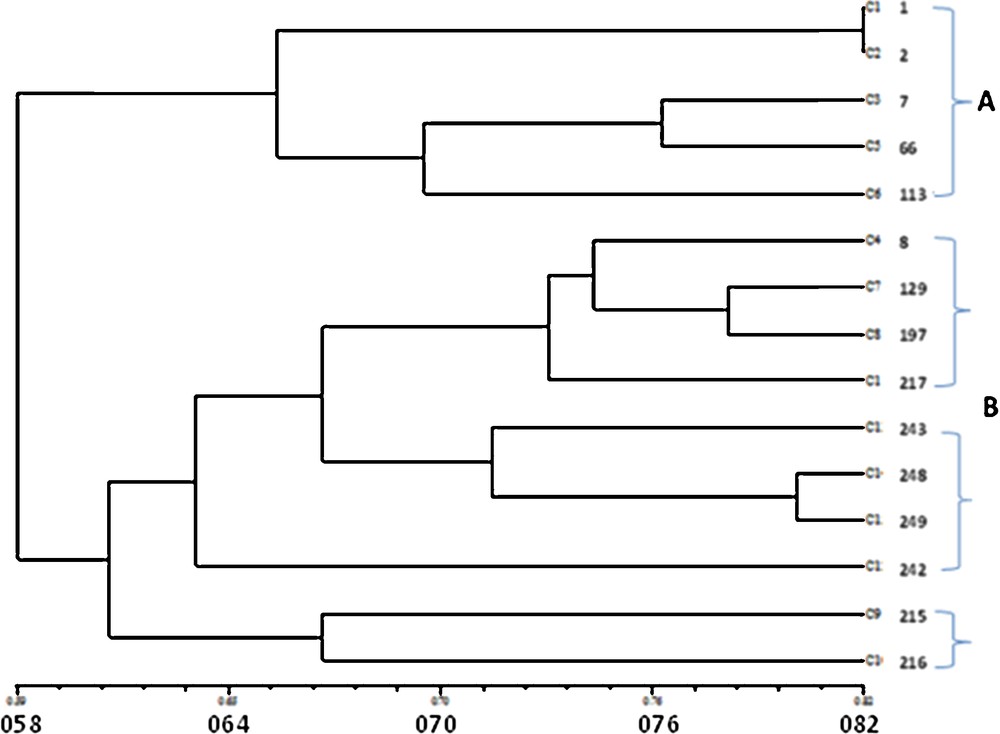
Dendrogram illustrating the genetic relationships among the 15 genotypes examined of sorghum based on ISSR polymorphism.
The data described above show that landraces are grouped based on grain color or geographic distribution. The genotypes in the southwest of Saudi Arabia (Jazan and Abha) were closer to each other than samples collected from Al-Baha further north or Al-Qassim in the central parts of Saudi Arabia. The ISSR analysis also differentiated landraces of white grains from landraces with dark grains. The differentiation of landraces of sorghum by molecular markers based on their geographic distribution was similarly reported in Ethiopian and Eritrean material [14]. In Zambia, most sorghum accessions were grouped according to their sites of collection based on SSR data, although some accessions that originated from the same site were grouped in different clusters [17]. Cluster analysis based on ISSR markers in cultivated sorghum differentiated resistance resources from susceptible control and hybrid parental lines. It was interesting to note that the susceptible hybrid parents and susceptible control were genetically similar, and the resistance sources were genetically a diverse group [21].
The present ISSR data generated by seven primers indicated a fairly high genetic diversity in the examined material and could be used for an efficient identification and detailed assessment of genetic diversity among sorghum landraces growing in Saudi Arabia and Yemen. This will help in the collection and authentication of the sorghum germplasm in the KACST gene bank. ISSR markers are of high value for sorghum germplasm characterization and conservation for future utilization. Consideration should also be given to other factors such as ecogeographic and ethnic differences when sampling sorghum genetic resources. However, the number of genotypes and the number of primers must be increased to better assess the genetic relationships among the landraces in more regions in the Arabian Peninsula.
Acknowledgement
The author is grateful to Prof. Turki A. Al Turk, for using his laboratory's facilities at KACST to perform the ISSR fingerprinting and to Prof. Abdelfattah Badr, Professor of Genetics and Plant Biosystematics, Helwan University, Egypt for advice on the preparation of the article and revising the manuscript.


