1 Introduction
Although relationships within the large and complex series of Atherinomorpha fishes (Teleostei) have been widely examined [1], there are still many uncertainties. Within this series, the order Atheriniformes (Table 1) comprises the respective suborders Atherinoidei (including the Atherinidae – Old World silversides) and Atherinopsoidei (including the Atherinopsidae – New World silversides). However, their separation into two differentiated suborders has not been still genetically verified. In addition, despite their significant biological and commercial value, the genetic relationships among these fishes remain largely unknown.
Specimens and abbreviation codes, location of collection sites, sample size, haplotypes and frequencies (in parenthesis) for each mitochondrial region.
| Haplotypes | ||||||||
| Species | Code | Locality | n | Phe + 12S rRNA | Cytb | COI | Thr + Pro + CR | Combined markers |
| Atherinopsoidei | ||||||||
| Atherinopsidae | ||||||||
| O. argentinensis | Oarg | Mar Chiquita lagoon, Argentina | 1 | 1(1) | 1(1) | 1(1) | 1(1) | 1(1) |
| Mar del Plata, Argentina | 2 | 1(2) | 1(2) | 2(1), 3(1) | 2(1), 3(1) | 2(1), 3(1) | ||
| O. smitti | Osmi | Mar del Plata, Argentina | 3 | 2(2), 3(1) | 1(3) | 4(2), 5(1) | 4(1), 5(1), 6(1) | 4(1), 5(1), 6(1) |
| O. incisa | Oin | Mar del Plata, Argentina | 3 | 4(1), 5(1), 6(1) | 1(3) | 6(1), 7(1), 8(1) | 7(1), 8(1), 9(1) | 7(1), 8(1), 9(1) |
| O. bonariensis | Obon | Gómez lagoon, Argentina | 3 | 7(2), 8(1) | 2(3) | 9(2), 10(1) | 10(1), 11(1), 12(1) | 10(1), 11(1), 12(1) |
| O. hatcheri | Ohat | Nahuel Huapi lake, Argentina | 1 | 9(1) | 3(1) | 11(1) | 13(1) | 13(1) |
| Atherinoidei | ||||||||
| Atherinidae | ||||||||
| A. hepsetus | Ath | Calella de Palafrugell, Spain | 4 | 10(4) | 4(4) | 12(1), 13(2), 14(1) | 14(1), 15(1), 16(1), 17(1) | 14(1), 15(1), 16(1), 17(1) |
| A. boyeri | Atb | Mar Menor coastal lagoon, Spain | 4 | 11(3), 12(1) | 5(3), 6(1) | 15(3), 16(1) | 18(1), 19(1), 20(2) | 18(1), 19(1), 20(2) |
| Melanoteniidae | ||||||||
| Melanotaenia sp | Mel | Unknown | 4 | 13(4) | 7(4) | 17(4) | 21(4) | 21(4) |
The genus Odontesthes, commonly known as “pejerreyes”, represents most of the New World silversides, being exclusive to South American waters (Fig. 1) [2]. Valued in fisheries and aquaculture, they have attracted sport fisheries as well [3]. Species of primary interest in the southwestern Atlantic include O. argentinensis Valenciennes, 1835, with commercial and sport fisheries in estuaries and coasts from Brazil, Uruguay and Argentina; O. bonariensis Valenciennes, 1835, a freshwater fish distributed in lakes and lagoons from Province of Buenos Aires (Argentina) and Rio Grande do Sul (Brazil); O. hatcheri Eigenmann, 1909, inhabiting Patagonian lakes and rivers of Argentina and Chile [2]; O. smitti Lahille, 1929, distributed along the Atlantic coast of Uruguay and Argentina and passing through Strait of Magellan to the Pacific coast of Chile [4,5]; and O. incisa Jenyns, 1841, distributed in the Atlantic coast from Dos Patos lagoon (Brazil) to Golfo Nuevo (Argentina) [5]. Molecular phylogenetic studies have been restricted to O. argentinensis (allozymes [6]; microsatellites and mtDNA control region [7]), where a species-wide morphological uniformity [2] contrasts with the incipient estuarine and coastal species suggested by these genetic studies. According to Froese [8], there is the need of deep revisions and identification catalogues of South American fishes, especially those from freshwater. In spite of this, few molecular studies of Odontesthes species exist and there are no studies on their molecular systematics.
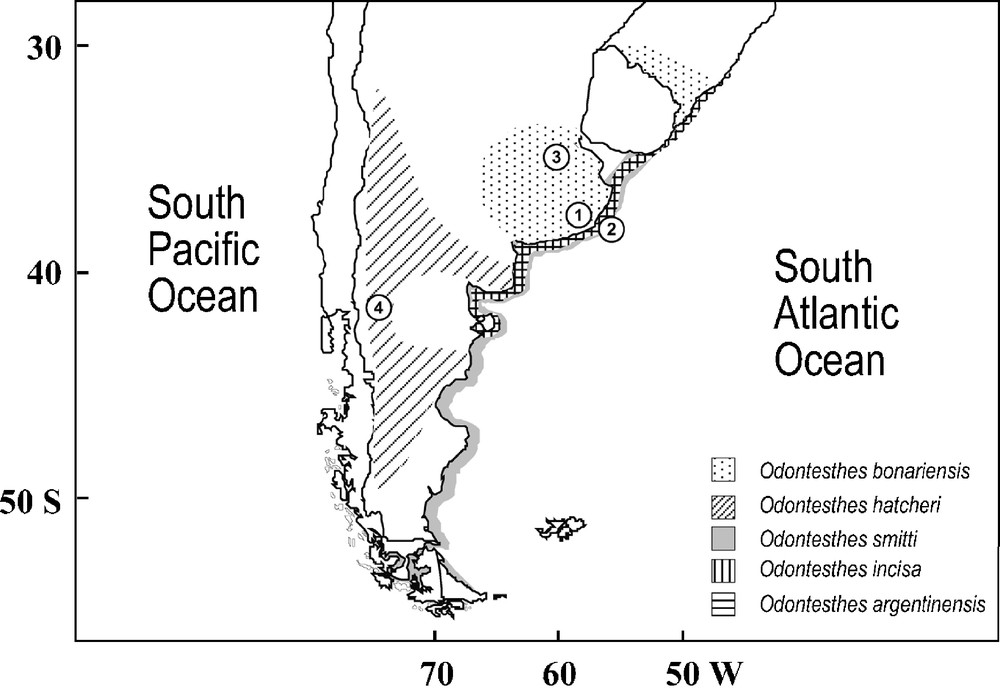
Distribution and sampling locations of Odontesthes species analyzed. 1: Mar Chiquita lagoon; 2: Mar del Plata; 3: Gómez lagoon; 4: Nahuel Huapi lake.
Within Atherinidae, genus Atherina is represented in Mediterranean waters by two species with commercial interest [9,10]: A. hepsetus Linnaeus, 1758, a typical marine silverside and Atherina boyeri Risso, 1810, which is distributed in lagoons, estuaries and marine environments. Systematic questions concern the relationships between these species, and the existence of a species complex within A. boyeri constituted by a marine non-punctuated form, a marine form with dark spots along the lateral line and a lagoon form [11–19].
As molecular markers, we used four regions of the mitochondrial genome to examine intra- and interspecific relationships within Odontesthes, providing a first molecular contribution to the understanding of their and Atherina phylogenetic relationships. We also clarify intergeneric relationships within Atheriniformes and report inter- and intrafamilial genetic distances for future taxonomical comparisons.
2 Material and methods
2.1 Sampling
Details of the 21 specimens collected for this study include individuals of two genera and seven species; Melanotaenia sp., belonging to the same suborder as Atherina genus (Atherinoidei), was also analyzed (Table 1). Morphological identification was based on Bauchot [9], Cousseau and Perrotta [5] and Dyer [2]. Muscle tissue was excised and conserved in 95% ethanol at the Laboratori d’Ictiologia Genètica collection.
2.2 PCR and sequencing
DNA isolation and polymerase chain reaction of phenylalanine transfer RNA (Phe), 12S rRNA, cytochrome b (cytb), cytochrome c oxidase subunit I (COI), threonine transfer RNA (Thr), proline transfer RNA (Pro) and control region (CR) were carried out in 50 μl reaction volume following Heras et al. [20]. The Phe and 12S rRNA region were amplified using the following pair sets of primers: L15927-Thr: 5′-AGA GCG TCG GTC TTG TAA TCC G-3′ [21] + 12ASR-H: 5′-ATA GTG GGG TAT CTA ATC CCA GTT-3′ [22] and L1091: 5′-CAA ACT GGG ATT AGA TAC CCC ACT AT-3′ [23] + H1478: 5′-TGA CTG CAG AGG GTG ACG GGC GGT GTG T-3′ [23] for Odontesthes spp., L15927-Thr + H1358-12S: 5′-CGA CGG CGG TAT ATA GGC-3′ [21] and L1091 + H1478 for Atherina spp. and DLHR: 5′-CAT CTG GTT CTT ACT TCA GG-3′, reverse of DLH, [24] + H1358-12S and L1091 + H1478 for Melanotaenia sp. For cytb amplifications, we used L14850-CYB: 5′-GCC TGA TGA AAC TTT GGC TC-3′ and H15560-CYB: 5′-TAG GCA AAT AGG AAG TAT CA-3′ [25] for all species. FishF2 5′-TCG ACT AAT CAT AAA GAT ATC GGC AC-3 and FishR1 5′-TAG ACT TCT GGG TGG CCA AAG AAT CA-3′ [26] were applied for COI PCRs for all species and finally, L15927-Thr and CSBDH: 5′-TGA ATT AGG AAC CAG ATG CCA G-3′ [27] for that of Thr, Pro and CR. Amplifications were verified on 1% agarose gel with ethidium bromide (0.5 mg ml−1) and cleaned in a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Little Chalfont, UK). Sequencing was performed by BigDye terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) according to manufacturer's instructions and fragments were read with an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems) at our laboratory. The nucleotide sequence of each gene in all species was verified from both strands. The primers (3.2 μM) used for forward and reverse strands sequencing were the same as for PCR.
2.3 Genetic analyses
Sequences were processed using SeqScape v2.5 (Applied Biosystems) and final alignments and sequence editing were carried out with BioEdit v7.0.4.1 [28]. DAMBE v4.5.61 [29] plots of the accumulated transitions and transversions vs Tamura and Nei [30] genetic distance were employed to test for nucleotide saturation. Tamura and Nei [30] mean distance values between species groups were obtained by MEGA v4 [31] for all mitochondrial regions separately. The partition homogeneity test (α = 0.05) was conducted with PAUP* v4.0b10 [32] to examine the homogeneity or incongruence between mitochondrial regions to be combined. Phylogenetic relationships were inferred by: neighbor-joining (NJ) analysis based on Tamura and Nei model employing MEGA and based on a maximum likelihood (ML) distance matrix employing PAUP*; maximum parsimony (MP) analysis and ML analysis both performed using PAUP* with an heuristic search and TBR branch-swapping algorithm with 10 random sequence addition; and lastly, Bayesian inference by MrBayes v3.1.2 [33]. Modeltest v3.7 [34] selected GTR + I + G as the best-fit model under Akaike information criterion for ML analyses and MrModeltest v2.2 [35] estimated GTR + I + G as the evolutionary model to run MrBayes program. Metropolis-coupled Markov chain Montecarlo (MCMC) analysis in MrBayes was achieved with four chains of 1 × 106 generations sampled every 100th and discarded 25% of samples as burn-in. A consensus tree with branch length and clade credibility (posterior probability) was generated with the 75% remaining samples. Robustness of trees was tested using bootstrap analysis [36] with 1000 replicates except for Bayesian analysis, which employed posterior probability for clade credibility. Unclear relationships inside Acanthopterygii regarding Atheriniformes were reported before [37] but available information indicate that Paracanthopterygii are their common ancestor, consequently, Gadus morhua (Paracanthopterygii - GenBank accession no. X99772) was used as outgroup species for all analyses.
3 Results
3.1 Haplotype diversity
Sequence alignment of Phe + 12S rRNA (908 bp), cytb (702 bp), COI (651 bp) and Thr + Pro + CR (533 bp) produced 13 (GenBank accession no. GQ352651–GQ352663), 7 (GenBank accession no. GQ352666–GQ352672), 17 (GenBank accession no. GQ352673–GQ352689) and 21 (GenBank accession nos. GQ352690–GQ352710) haplotypes, respectively (Table 1). Note that O. argentinensis (GQ352664), O. smitti (GQ352665) and O. incisa (GQ352666) shared haplotype 1 for cytb indicating the low evolutionary signal of this molecular marker for discerning among these taxa and suggesting its elimination for latter analyses. In contrast, no shared haplotypes between species were detected for the rest of mitochondrial genes. Furthermore, no clear association between haplotypes and environmental distribution for O. argentinensis was observed because of shared haplotypes detected between individuals from Mar Chiquita lagoon (estuarine) and Mar del Plata (open sea) in Phe + 12S rRNA and cytb (Table 1).
3.2 Genetic divergence
No saturation was detected for used markers (data not shown). As expected, the pairwise Tamura and Nei distance matrix generated from each dataset (Table 2) identified much higher distances for intergeneric comparisons. All genera appeared almost equally separated among them for Phe + 12SrRNA and COI, but for Thr + Pro + CR, the proximity of Atherina and Melanotaenia versus Odontesthes was apparent. The lowest divergence values were those between O. argentinensis–O. bonariensis and O. smitti–O. hatcheri for all molecular markers, indicating close genetic relationship between these paired taxa (Table 2).
Tamura and Nei [30] mean genetic distances and standard error for each sample and three mitochondrial markers. Species codes are as in Table 1.
| Phe + 12S rRNA | COI | Thr + Pro + CR | |
| Within Odontesthes | |||
| Oarg | 0 | 0.0031 ± 0.0017 | 0.0116 ± 0.0042 |
| Osmi | 0.0011 ± 0.0011 | 0.0015 ± 0.0014 | 0.0131 ± 0.0042 |
| Oin | 0.0022 ± 0.0013 | 0.0083 ± 0.0029 | 0.0087 ± 0.0034 |
| Obon | 0.0011 ± 0.0011 | 0.0031 ± 0.0020 | 0.0237 ± 0.0061 |
| Between Odontesthes | |||
| Oarg-Osmi | 0.0119 ± 0.0037 | 0.0302 ± 0.0067 | 0.0693 ± 0.0119 |
| Oarg-Oin | 0.0105 ± 0.0033 | 0.0516 ± 0.0083 | 0.0654 ± 0.0112 |
| Oarg-Obon | 0.0017 ± 0.0012 | 0.0046 ± 0.0020 | 0.0253 ± 0.0054 |
| Oarg-Ohat | 0.0113 ± 0.0037 | 0.0294 ± 0.0065 | 0.0680 ± 0.0119 |
| Osmi-Oin | 0.0058 ± 0.0023 | 0.0405 ± 0.0078 | 0.0540 ± 0.0101 |
| Osmi-Obon | 0.0136 ± 0.0039 | 0.0291 ± 0.0066 | 0.0703 ± 0.0111 |
| Osmi-Ohat | 0.0039 ± 0.0020 | 0.0101 ± 0.0039 | 0.0204 ± 0.0054 |
| Oin-Obon | 0.0100 ± 0.0032 | 0.0516 ± 0.0083 | 0.0629 ± 0.0102 |
| Oin-Ohat | 0.0075 ± 0.0027 | 0.0439 ± 0.0081 | 0.0546 ± 0.0106 |
| Obon-Ohat | 0.0130 ± 0.0040 | 0.0283 ± 0.0066 | 0.0704 ± 0.0115 |
| Within Atherina | |||
| Ath | 0 | 0.0062 ± 0.0024 | 0.0084 ± 0.0028 |
| Atb | 0.0022 ± 0.0016 | 0.0015 ± 0.0015 | 0.0054 ± 0.0026 |
| Between Atherina | |||
| Atb-Ath | 0.0479 ± 0.0076 | 0.1297 ± 0.0151 | 0.2162 ± 0.0226 |
| Odontesthes vs Atherina | |||
| Oarg-Atb | 0.1643 ± 0.0150 | 0.2336 ± 0.0224 | 0.5740 ± 0.0522 |
| Oarg-Ath | 0.1685 ± 0.0150 | 0.2253 ± 0.0217 | 0.5156 ± 0.0455 |
| Osmi-Atb | 0.1602 ± 0.0150 | 0.2173 ± 0.0212 | 0.5937 ± 0.0542 |
| Osmi-Ath | 0.1643 ± 0.0151 | 0.2153 ± 0.0205 | 0.5633 ± 0.0488 |
| Oin-Atb | 0.1636 ± 0.0149 | 0.2185 ± 0.0211 | 0.6114 ± 0.0561 |
| Oin-Ath | 0.1677 ± 0.0150 | 0.2233 ± 0.0213 | 0.5664 ± 0.0492 |
| Obon-Atb | 0.1652 ± 0.0152 | 0.2302 ± 0.0222 | 0.5775 ± 0.0521 |
| Obon-Ath | 0.1694 ± 0.0152 | 0.2221 ± 0.0213 | 0.5248 ± 0.0463 |
| Ohat-Atb | 0.1637 ± 0.0150 | 0.2253 ± 0.0221 | 0.5977 ± 0.0543 |
| Ohat-Ath | 0.1678 ± 0.0151 | 0.2174 ± 0.0206 | 0.5731 ± 0.0503 |
| Odontesthes vs Melanotaenia | |||
| Oarg-Mel | 0.1714 ± 0.0163 | 0.2219 ± 0.0215 | 0.5595 ± 0.0496 |
| Osmi-Mel | 0.1706 ± 0.0161 | 0.2234 ± 0.0210 | 0.5972 ± 0.0538 |
| Oin-Mel | 0.1708 ± 0.0160 | 0.2207 ± 0.0201 | 0.6093 ± 0.0548 |
| Obon-Mel | 0.1737 ± 0.0162 | 0.2206 ± 0.0211 | 0.5576 ± 0.0486 |
| Ohat-Mel | 0.1726 ± 0.0162 | 0.2199 ± 0.0206 | 0.5904 ± 0.0525 |
| Atherina vs Melanotaenia | |||
| Atb-Mel | 0.1586 ± 0.0149 | 0.2096 ± 0.0206 | 0.4240 ± 0.0369 |
| Ath-Mel | 0.1772 ± 0.0160 | 0.2068 ± 0.0207 | 0.4040 ± 0.0345 |
| Odontesthes vs Atherina vs Melanotaenia | |||
| Odontesthes spp-Atherina spp | 0.1660 ± 0.0147 | 0.2237 ± 0.0193 | 0.5706 ± 0.0463 |
| Odontesthes spp-Melanotaenia sp | 0.1717 ± 0.0163 | 0.2214 ± 0.0202 | 0.5816 ± 0.0506 |
| Atherina spp-Melanotaenia sp | 0.1710 ± 0.0150 | 0.2085 ± 0.0191 | 0.4155 ± 0.0328 |
3.3 Phylogenetic analysis
Partition homogeneity test on 2794 bp excluded cytb (P < 0.0001) as noted above (Table 1), establishing the congruence of combining Phe + 12S rRNA, COI and Thr + Pro + CR, and generating a consensus alignment of 2092 bp (P = 0.7900). Phylogenetic analysis generated similar topologies of the trees indicating two major different lineages or phylogroups (Fig. 2). One lineage comprised all Odontesthes spp. haplotypes (100% robustness) clearly supporting a monophyletic group. It is noteworthy that haplotypes of O. argentinensis from Mar del Plata open sea (Oarg2 and Oarg3) clustered together. In all the analyses, O. argentinensis and O. bonariensis had minima distance values and shared a common ancestor (99% of bootstrap support). Similarly, O. smitti and O. hatcheri were closely related (99% of bootstrap support) and grouped with O. incisa haplotypes (81% of bootstrap support). A second lineage was formed by A. boyeri and A. hepsetus (100% robustness) which strongly clustered with Melanotaenia sp (100% of bootstrap support).
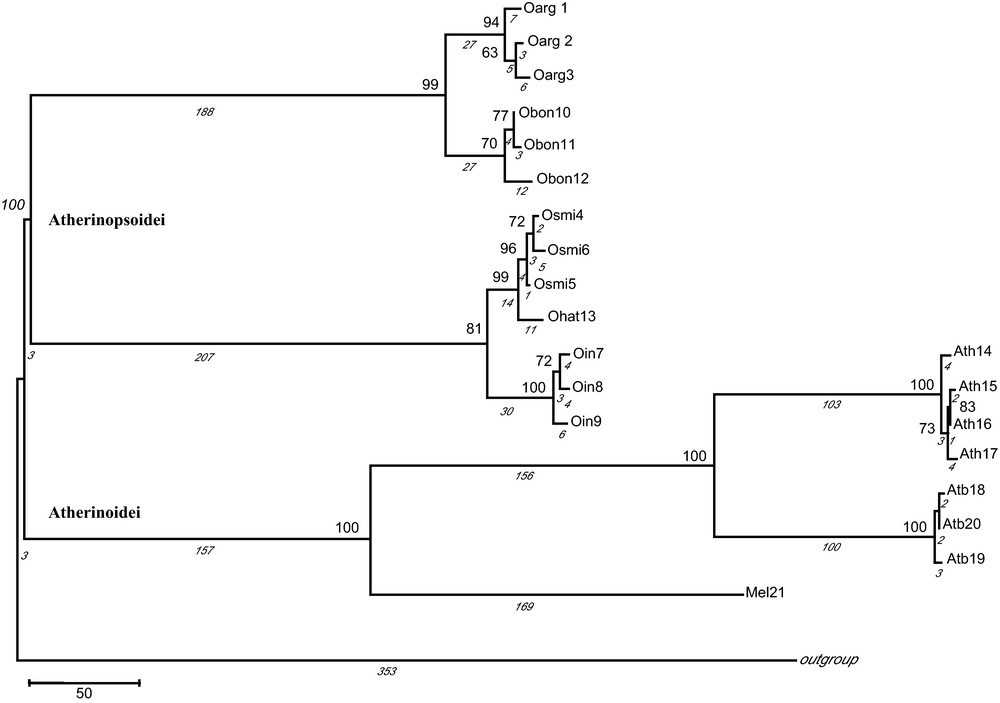
Maximum parsimony tree with combined markers based on 573 parsimony informative sites. The numbers in nodes indicate ≥ 60 bootstrap value. The values below branches (in italics) indicate the number of mutational steps (1641 in total). Haplotype codes are as in Table 1. Gadus morhua was used as outgroup species.
We constructed additional phylogenies on Atherina spp., incorporating comparable data of 12S rRNA (Fig. 3A) (42 haplotypes from 165 sequences) and control region (Fig. 3B) (300 haplotypes from 453 sequences) from GenBank [13,16,17,38] with those of our study. In this analysis, our A. hepsetus haplotypes clustered with those from previous authors and grouped with A. presbyter. In this analysis, our A. boyeri haplotypes matched the lagoon-estuarine type and was clearly differentiated from the marine types punctuated (P) and non-punctuated (NP) that emerged into two diverged lineages.
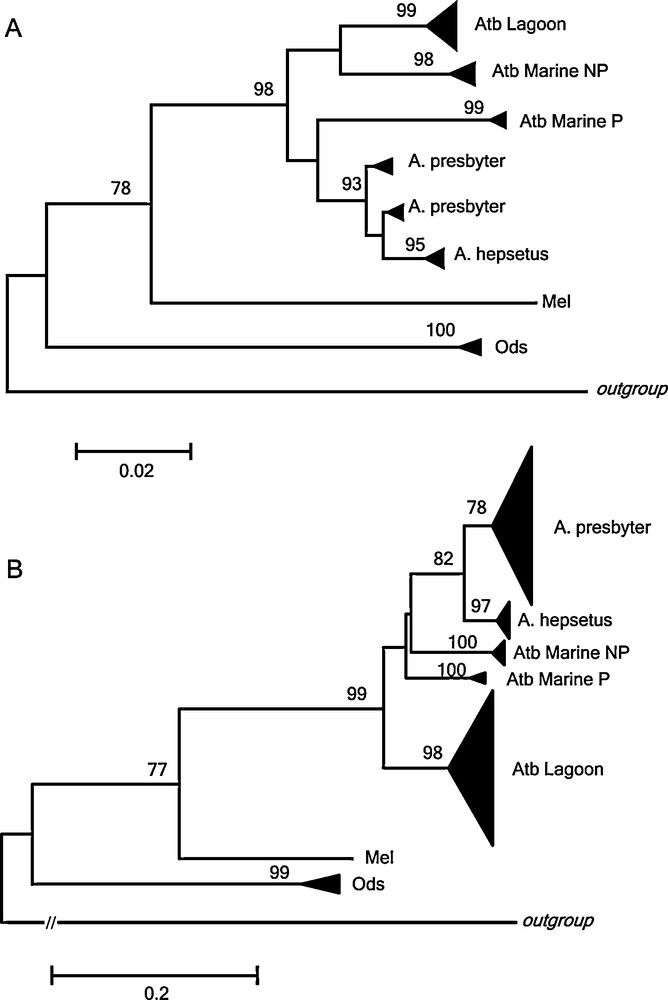
Comparative study in Atherina spp. A: Neighbor-joining tree of 12S rRNA (351 bp) based on Tamura and Nei [30] distances using 31 sequences from this study, and 165 sequences from other authors. B: Neighbor-joining tree of control region (381 bp) based on Tamura and Nei [30] distances using 31 sequences from this study and 453 sequences from other authors. The numbers on nodes indicate ≥ 60 bootstrap values. Triangle sizes are proportional to the number of haplotypes present in the cluster. NP = non-punctuated; P = punctuated. Gadus morhua was used as outgroup species. Masquer
Comparative study in Atherina spp. A: Neighbor-joining tree of 12S rRNA (351 bp) based on Tamura and Nei [30] distances using 31 sequences from this study, and 165 sequences from other authors. B: Neighbor-joining tree of control region (381 bp) ... Lire la suite
4 Discussion
4.1 Odontesthes phylogeny
In corroborating the monophyly of the genus Odontesthes analysing five species from marine and freshwater environments, our results support the morphological analyses revised by Dyer [2] based on anatomical characters. For all mitochondrial regions, genetic distances between Odontesthes species were relatively low in comparison with those relative to Atherina species, indicative that genus Odontesthes radiated as recently as cichlid fishes in Lake Victoria [39]. Genetic distances in COI fit the most frequent value between congeneric fishes previously reported by Ward ([40]; D ≈ 0.035). Likewise, the sympatric distribution of the marine species O. argentinensis, O. smitti and O. incisa, could be explained as a secondary contact after allopatric speciation as indicated by the splitting in three different nodes (Fig. 2).
Within the Odontesthes cluster (Fig. 2), O. argentinensis from Mar del Plata open sea (Oarg2 and Oarg3) grouped together and split off from that of Mar Chiquita estuarine lagoon (Oarg1), suggesting the occurrence of a barrier to gene flow [41] between these two environments. These findings are consistent with genetic studies using allozymes [6] and microsatellites and mitochondrial control region [42] and morphological landmarks [43], each indicating differentiated populations or even incipient species of O. argentinensis from marine and estuarine environments along Brazilian and Uruguayan coasts, respectively. Moreover, spawning grounds and timing in the two forms of O. argentinensis are different, as well as their habitat preference. Temporal and spatial separation of their life cycle implying an ecological divergence [6] may contribute to create one or several mechanisms producing complete reproductive isolation between them [44]. Moreover, there is no geographical barrier between Mar Chiquita lagoon and Mar del Plata open sea in Argentina separating the lagoon-estuarine form from the marine one, like in the Brazilian area studied by [6]. However, our reduced number of individuals and haplotypes and the low genetic distances, characteristic of intraspecific levels, do not allow us to establish two valid species. Nonetheless, bottlenecks or founder effects may determine the differentiation in O. argentinensis populations of brackish environments coming from an ancestral marine population [6], provoking a feasible beginning of ecological speciation [45]. We consider O. argentinensis as an interesting model for divergent adaptive selection studies associated to reproductive isolation mechanisms, consequently, the preservation of the marine and estuarine-lagoon populations must be taken into account for biodiversity conservation policies in the Mar Chiquita lagoon, designated a UNESCO World Biosphere reserve in 1996 [46].
Genetic distances detected between O. bonariensis (freshwater fish) and O. argentinensis for all molecular markers were within the range of Odontesthes intraspecific levels (Table 2). Moreover, O. bonariensis and O. argentinensis comprised a common lineage in all phylogenetical analyses (Fig. 2) consistent with their shared morphological characters [47]. In addition, Tombari et al. [48] pointed out that O. bonariensis and O. argentinensis were difficult to distinguish morphologically with no significant morphometric differentiation. Moreover, Tejedor [49] observed natural interbreeding between these two taxa in the La Salada Grande lagoon (General Madariaga, Argentina). O. bonariensis, a freshwater fish would be the product of the speciation of a common marine ancestor shared with O. argentinensis [49] as well as with O. perugiae species complex [7]. Experimental works that could demonstrate speciation at present time in nature had been considered unrealizable due to the assumption of a long time of isolation requirements. However, Hendry et al. [50] proved how the reproductive isolation could evolve in a fast way when new and different habitats were colonized. In that work, reproductive isolation was detected at only after 13 generations, representing a short evolution period of 56 years, between two populations of Oncorhynchus nerka coming from a common freshwater reproductive source that was introduced in different environments (coastal marine and river). Silversides exhibit a great ability for invading and speciating in vacant niches [51] like the Laguna de Gómez, where O. bonariensis lives, that have a connection towards Atlantic Sea through Río Salado, in front of Argentinean coastal waters where O. argentinensis occurs. Therefore, the high genetic variability of the Odontesthes marine-brackish ancestor would have preadapted them to colonize and speciate rapidly into new habitats [45,52]. Colonization of new estuarine habitats, with very unstable physical-chemical conditions and biological factors, would favour a fast adaptation and reproductive isolation by means of natural selection [53].
Similarly, our analyses revealed a close relationship between O. smitti (marine) and O. hatcheri (freshwater) (Table 2; Fig. 2), both joining O. incisa (marine) in a common cluster (Fig. 2), although Dyer [2] argued that O. hatcheri was the sister group for the entire genus Odontesthes. Despite the occurrence of spontaneous hybridization cases in captivity suggesting the proximity of O. bonariensis and O. hatcheri, both freshwater fishes were easily discriminated with all mitochondrial genes used in this study as well as in a previous study with RFLPs [54,55].
According to Bamber and Henderson [51], most atherinid fishes including genus Odontesthes from Patagonia, where O. hatcheri occurs, had mainly originated from marine immigrants. A rapid uplift of the Andes is speculated to have occurred at later Miocene (10–5 mya) [56] and until Pleistocene-Holocene (1.8 mya–10,000 ya) there was a mass of water at the continental platform due to marine transgressions (Paranense Sea, Patagonian Sea) [47,57]. Thus, O. hatcheri may have diverged just before the sea level descended at the one currently known in that area. In addition, phylogenetic analyses suggest a marine–freshwater pairing pattern in Odontesthes and an expected freshwater reciprocal fish for O. incisa, a hypothesis awaiting to be verified when the complete genus will be analyzed. Therefore, the vicariant segregation of a marine and freshwater species could be a result of both tectonic plates and glacial intervals provoking fluctuations in the sea level that depicted the major part of the variability at marine species from the Atlantic Ocean [58]. As was suggested by O. perugiae complex [45], change of the sea level during Pleistocene-Holocene could have originated the lineages derived from the sea in South America matching with the recent radiation of Odontesthes [59]. Given that vicariant patterns of speciation in marine fishes are difficult to detect due to insufficient phylogenetic analyses [60], Odontesthes phylogeny offers a great opportunity to study biogeographic processes in South America [61].
4.2 Atherina phylogeny
Recently A. boyeri as a single taxon has been questioned and a complex of forms has been proposed including: (1) two types, marine and lagoon [11–13,18] and (2a) three species, A. boyeri (marine non-punctuated), A. punctata (marine punctuated) and A. lagunae (lagoon) [14,15,19]; (2b) three species, marine non-punctuated form, marine punctuated form and A. boyeri (lagoon form) [17]. In the recent revision by Kottelat and Freyhof [62], the individuals named as A. lagunae by Trabelsi et al. [14,15] were in fact A. boyeri based on morphological diagnostic characters that were consistent with Francisco et al. [17] conclusions. In addition, A. punctata is an unavailable name due to its homonymy with A. punctata Bennet, 1833 and thus, the 21 available synonyms of A. boyeri should be considered and examined before naming this taxon [62,63]. To verify these proposals, we calculated mean Tamura and Nei [30] genetic distances in 12S rRNA (351 bp, 42 haplotypes) and control region (381 bp, 300 haplotypes) among all Atherina species and types to determine their degree of genetic differentiation. Genetic distances between the different types of A. boyeri (lagoon - marine punctuated were D = 0.0713 ± 0.0143, D = 0.2028 ± 0.0243; lagoon - marine non-punctuated were D = 0.0457 ± 0.0106, D = 0.1996 ± 0.0240; marine punctuated - marine non-punctuated were D = 0.0527 ± 0.0121, D = 0.1904 ± 0.0233), are noticeable higher than those detected between two recognized species of genus Atherina, A. hepsetus and A. presbyter (D = 0.0179 ± 0.0057, D = 0.1133 ± 0.0164). Consequently, given that species status of A. hepsetus and A. presbyter has not been questioned, these results together with the three phylogroups detected (Fig. 3) support the three different species associated with lagoon and marine environments of A. boyeri previously proposed constituted by: 1) the marine form without the presence of spots; 2) the marine form with dark spots along the lateral line, restricted to western Mediterranean Sea [19]; and 3) the brackish form from estuaries and lagoons. In this way, isolation that had operated over populations exploiting different niches [64] or environments with different salinities [52] would support the occurrence of ecological speciation. Thanks to silverside plasticity, that would imply a selection of generalist genotypes to confront a wide range of conditions [42], the ancestor of A. boyeri could have adapted to radiate repeatedly across vacant habitats, thus colonizing, estuaries and lagoons and acquiring also the ability to invade freshwater biotopes [51]. Thus, this specific variation (meristics, size, particular morphological structures, etc.) of some species as a result of a response to the ambience [52] due to both fluctuations of the Mediterranean Sea levels during Pleistocene age and the Messinian crisis [17] probably originated the species complex in A. boyeri.
4.3 Atheriniformes phylogeny
Genetic distances for COI between the three families analyzed, Atherinopsidae (Odontesthes), Atherinidae (Atherina) and Melanotaeniidae (Melanotaenia) (D = 0.2085–0.2237; Table 2) fit into the mean value between families of the same order of fishes (D = 0.2256; [40]). In addition, Atherinoidei was stated as a monophyletic group (Fig. 2). Nelson [65] included Atherinidae in the suborder Atherinoidei with two recognized groups, Old World and New World silversides as subfamilies. More recently, Nelson [1] revised the classification in the light of recent morphoanatomical systematic works [47,66], where: (1) Old World silversides are retained in the Atherinidae family in suborder Atherinoidei (Table 1); and (2) the family Atherinopsidae was created for New World silversides within a new suborder Atherinopsoidei (Table 1). Our results support the new taxonomical category assignments showing the closeness, both in the means of phylogenetic analyses and genetic distances, of Atherina and Melanotaenia (Atherinoidei), compared to Odontesthes (Atherinopsoidei) (Table 2; Fig. 2).
5 Conclusions
- 1 The close genetic relationships between Odontesthes species were indicative that this genus has radiated in relatively recent time;
- 2 The status of A. boyeri needs to be revised and three species are corroborated corresponding to a marine non-punctuated form, a marine punctuated form and a brackish form;
- 3 Odontesthes and Atherina are recognized as monophyletic genera;
- 4 The monophyly of suborders Atherinopsoidei and Atherinoidei is recognized.
As molecular data accumulate, more refined phylogenetic analyses will became possible. Meanwhile, our genetic data caste light on some taxonomical aspects of Atheriniformes. However, the whole Odontesthes phylogeny based on molecular data still remains unknown and further studies including other species of Odontesthes are required. In addition, we recommend studies on reproductive biology and population genetics within O. argentinensis and between O. argentinenesis–O. bonariensis and O. smitti–O. hatcheri in order to estimate interbreeding levels, and to corroborate or reject the incipient marine–freshwater dichotomy suggested in this work. Odontesthes and Atherina may represent geographically replicated ideal models to study genetic adaptation and speciation of marine fish to brackish and freshwater habitats.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The participation of Patricia Noguera, Juan Roldán, Martí Cortey, María Berta Cousseau, Mariano González-Castro, Mercedes González and Asunción Andreu made the collections possible. We would like to thank Fred Utter for helpful comments on the manuscript and corrections of the English style. Funding was provided to SHM by BRAE predoctoral fellowship and to MIR by 910305 grant from Universitat de Girona.


