1 Introduction
Transposable elements (TEs) are defined as DNA sequences that can move/transpose to new sites in the genome of a single cell, either by themselves or by hijacking active TE replication machinery. Yeasts are fungi able to multiply indefinitely as unicellular organisms. Their genomes contain several TE families that have already been described, belonging to two main classes: Class-I elements, also called retrotransposons, transpose using a copy-and-paste mechanism with an RNA as the intermediate, and Class-II elements or DNA transposons can move using a cut-and-paste mechanism with a DNA intermediate [1]. Class-I elements are often found in yeast genomes and were described several decades ago, whereas Class-II elements have only been found in a very limited number of species and were identified more recently. It is the mechanism of transposition that differentiates TEs from the other types of mobile genetic elements found in yeast genomes, including endonuclease-coding group-I introns [2,3], inteins [4], and mating type genes [5,6]. Only TEs present in yeast nuclear genomes will be considered in this review; Class-I reverse transcriptase-coding group-II introns, mobile elements present in some yeast mitochondrial genomes [7–9], will not be addressed here.
This short review surveys the current status of research on yeast TEs. This work has been facilitated by the takeoff of powerful high-throughput methods including microarray hybridization, whole genome sequencing and next-generation sequencing (NGS). We will focus on genomic variability associated with TEs and, in particular, the following issues will be considered: (i) state of the art of TE sampling across yeast species (full-length TEs as well as degenerate copies and relics will be considered); (ii) investigations of intraspecific diversity of TE content; (iii) effects of TEs on genome stability and on gene expression; and (iv) TE–host interactions.
2 Transposable elements genomic contents: interspecies diversity
All yeast species described to date belong to the Ascomycota and Basidiomycota. They mostly have small and compact genomes (between 10 and 20 Mb) with low TE contents (less than 5%) [10,11]. By contrast, some filamentous Ascomycota and Basidiomycota have much larger genomes with a high proportion of TEs, for instance Laccaria bicolor (65 Mb genome with 21% TE [12]), Tuber melanosporum (125 Mb with 58% TE [13]) and Blumeria graminis (118 Mb with 70% TE, www.blugen.org). The first TEs identified in yeasts were Class-I Ty elements bearing Long Terminal Repeats (LTR) [14]. The TEs Ty1 to Ty5 have been extensively studied in the model yeast Saccharomyces cerevisiae since the 1980s. LTR retrotransposons were then characterized in Schizosaccharomyces pombe [15] and in a few Saccharomycotina yeasts, particularly Candida albicans [16] and Yarrowia lipolytica [17], and the first non-LTR yeast retrotransposon was found in C. albicans [18] as was the first Class-II element [19]. The recent genome sequencing projects have led to the past ten years being highly productive for the discovery and description of new yeast TEs. Because of their repetitive nature and length exceeding that of a sequence read, TEs represent a challenge for assembling genome sequences. Indeed, manual finishing steps were required to resolve the structure of loci containing TEs, during genome sequencing by ‘Sanger’ technologies. For example, some of the TEs revealed by the Génolevures projects [20–24] have been individually reamplified by PCR from clones of DNA libraries or from genomic DNA, to confirm their presence at a given locus and to ensure high quality assembly of the respective genomes. With new sequencing technologies, mate-pair sequencing of inserts longer than TEs is required to resolve gaps in sequence assemblies. It has to be emphasized that even when the sequence data are available, identifying TEs in yeast genomes remains a painstaking task because of their low copy number, which hinders efficient detection by the current bioinformatic pipelines developed for repeated sequence analyses [25]. Their detection therefore depends heavily on alignments against known TE sequences or on manual scrutiny of particular regions believed to contain TEs. For instance, in S. cerevisiae Ty1, Ty2, Ty3 and Ty4 elements insert preferentially in clusters upstream from genes transcribed by RNA polymerase III, i.e. tRNA genes, 5S rRNA genes and a few noncoding RNA genes [26–28], whereas in S. pombe, Tf1 and Tf2 integrate with a strong preference for RNA polymerase II-transcribed gene promoters [29,30]. Ty5 insertions have been found within heterochromatin regions present at the silent cassettes of the mating-type loci and at telomeres as a consequence of an interaction between Ty5 integrase and the protein Sir4 [31,32]. Tdh5 from Debaryomyces hansenii and Tps5 from Pichia stipitis were found as one cluster per chromosome, at a site that is possibly the centromeric region [33,34]. It may be feasible to automate the manual detection based on these considerations by further developing bioinformatics tools.
The diversity of TEs identified in yeast genomes is illustrated in Fig. 1. Despite the prevalence of LTR retrotransposons, representatives of a large spectrum of the eukaryotic TEs are found in yeasts and, in particular, Long Interspersed Repetitive Elements (LINE) and several Class-II element families are also present in yeast genomes. However, yeast species display substantial differences in their TE contents with patchy distributions of TE families and variations in copy numbers. Some species can be qualified as TE ‘reservoirs’ either because they contain potentially active TEs belonging to diverse classes and families (C. albicans, Cryptococcus neoformans, Y. lipolytica) or because they present a lower diversity of TE families but high copy numbers (S. cerevisiae). Sequenced strains of other species are almost empty of TEs either because all the TEs detected are not active (degenerate or relic copies) (Eremothecium gossypii and Zygosaccharomyces rouxii) or because no trace of TE has been detected so far (Pichia sorbitophila, personal communication). From a biological point of view, it is of great importance to distinguish, among inactive copies of TE, the variously degenerated TE relics (for instance solo-LTRs which are remnants of former complete TEs) from full-length mutated copies (for instance all the copies of Kluyveromyces lactis Tkl1). Indeed, recombination between two mutated copies can regenerate potentially active copies, as has been described for Ty1 [35], and can result from efficient recombination during cDNA synthesis in heterozygous Virus Like Particles (VLP), by template switching of the reverse transcriptase.
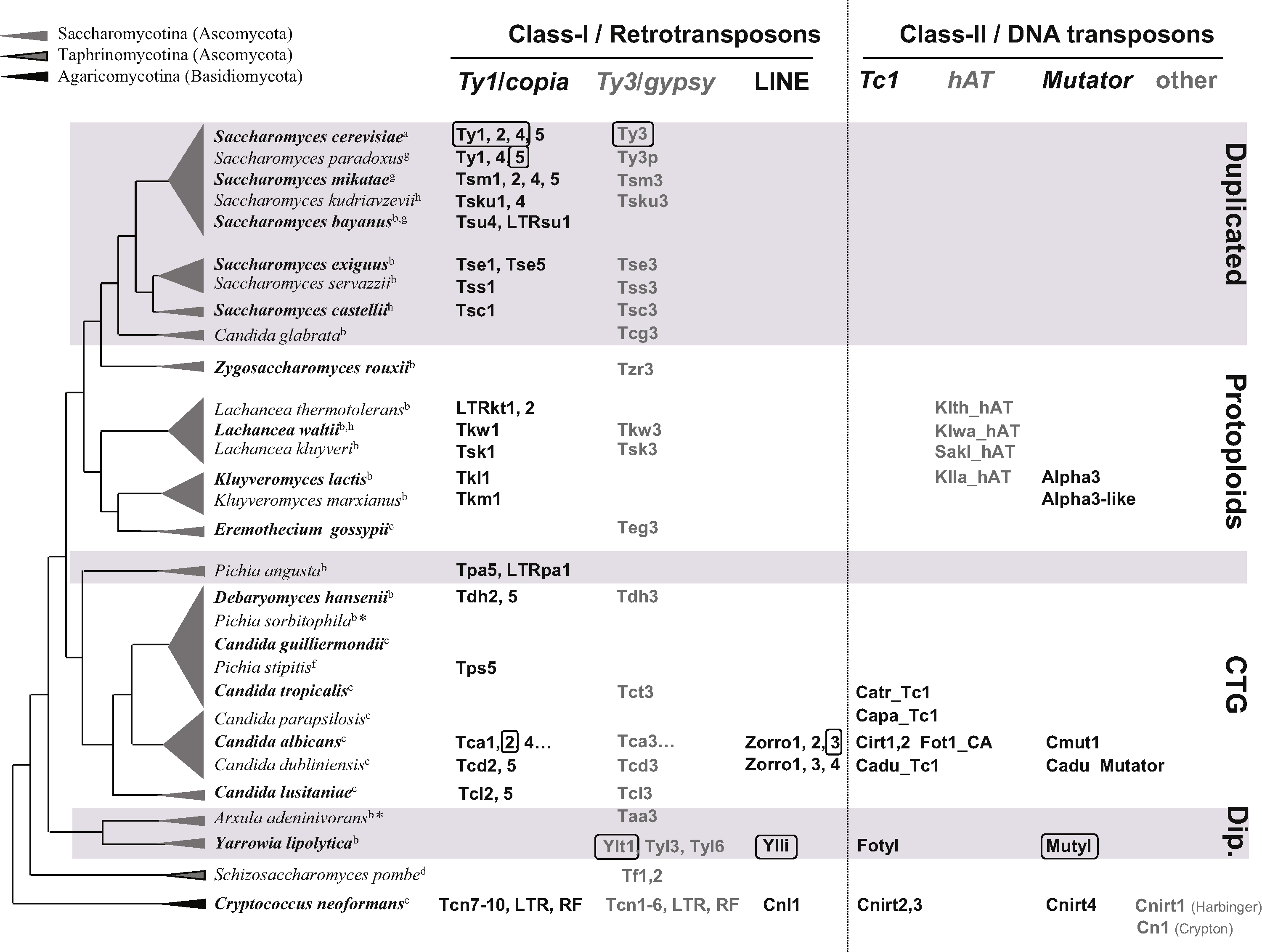
Overview of the Transposable element (TE) diversity in yeast species. The figure summarizes the list of the Ascomycota and Basidiomycota yeast species in which TE contents have been investigated. The tree topology is adapted from ref. [107], the branch lengths are arbitrary. Major yeast families in Saccharomycotina are shown with white or gray backgrounds (Dip.: Dipodascaceae). For each species, the presence and the respective names of TEs of the major families of Class-I and Class-II transposons are indicated, for a better legibility, yeast TE names were not written in italic. The Long Terminal Repeats (LTR)-retrotransposon nomenclature is based on ref. [20] (except for Ty elements). TEs for which data are available evidencing effective transposition activity are indicated by boxes. Fig. 1 includes the Kluyveromyces lactis Alpha3 transposable element presenting a transposase activity required for Mating-type switching in this species and representing an interesting case of TE domestication [91]. Some families are only represented by TE relics (LTR and RF, RF: Retrotransposon Fragment). Genomic sequences and annotations of yeast species are available at: ahttp://www.yeastgenome.org/; bhttp://www.genolevures.org, (*: submitted); chttp://www.broadinstitute.org/annotation/fungi/fgi/; dhttp://old.genedb.org/genedb/pombe/. ehttp://agd.vital-it.ch/Ashbya_gossypii; fhttp://genome.jgi-psf.org/Picst3/; ghttp://www.broadinstitute.org/annotation/fungi/comp_yeasts/; hhttp://wolfe.gen.tcd.ie/ygob.
Several comprehensive hypotheses have been suggested to explain the distribution of yeast TEs:
- • The phylogeny of copia-like LTR transposons in Saccharomycetaceae suggests that they are all descended from a common ancestral element that evolved in the different lineages by sequential acquisition of novel structural features representing essential steps in LTR retrotransposon replication. These features provide them with selective advantages, for example, the different frameshift mechanisms regulating the Gag/Gag-Pol ratio and the primer binding sites complementary to different tRNAs that initiate the first-strand reverse transcription [20]. For instance, Ty1-like elements (Tse1 to Tkm1) characterized by a +1 frameshift mechanism and using tRNAiMet as primer may have emerged in yeast species after the divergence from the CTG lineage (Fig. 1).
- • Some elements may have been specifically acquired by some clades, probably by horizontal transfer, as illustrated by the hAT-like elements in Lachancea species [24] and by the Tc1-Mariner and Mutator elements in C. albicans and Yarrowia species [23] and, as is suspected, by Tsk1 in Lachancea kluyveri [20,36].
- • The variability in TE content may also depend on whether some lifestyles may favor TE reinvasion and expansion/retention or may alternatively accelerate TE loss/elimination. For example, the ‘TE reservoir’ species (C. albicans, C. dubliniensis, C. neoformans) are pathogenic for humans and TE abundance seems to be a signature for micro-organisms that have undergone an adaptative evolutionary pathway; examples include the destructive potato pathogen oomycete Phytophthora infestans [37], the cereal pathogens belonging to Blumeria species [38] and the fungal symbionts L. bicolor and T. melanosporum [12,13]. However, the fact that the human pathogen C. glabrata and the osmotic resistant species P. sorbitophila lack TEs indicates that TE abundance is not a general rule for such organisms.
- • The substantial differences in TE content between yeast species and the fact that some TEs are present at very low copy number or only as degenerate copies raise the issue of whether these observations represent general traits shared by the species or specific features acquired by particular strains. This encourages investigations into the extent of TE diversity between several strains and even isolates of a given species.
3 Genomic Transposable elements content: intraspecific diversity
TE insertions vary in type and copy number between species. In addition, studies with numerous organisms including yeasts have demonstrated that TEs also vary in content and location between individuals/isolates of the same species. TE-related polymorphism provides the richest sources of genetic variability, along with single nucleotide polymorphism (SNP) and subtelomeric region variability [39]. Such polymorphism is long-established in S. cerevisiae [40,41] and is exploited for genotyping by interdelta PCR [42]. However, in contrast to SNP mapping, establishing the precise TE landscape of a genome remains complex.
Southern-based chromosome hybridization and comparative genomic hybridization have been used to investigate both the TE type and copy number, but such experiments do not provide their exact positions. These studies have demonstrated the high variability in Ty element contents between S. cerevisiae strains (see [43,44], for a few examples), S. paradoxus isolates [31,44] and a collection of strains from different Saccharomyces sensu stricto species [45]. The diversity of the Tca2 retrotransposon in C. albicans strains [46] and the diversity of the Ylli LINE retrotransposon, Mutyl Class-II element and Tyl6 LTR-retrotransposon have also been investigated in Y. lipolytica lineages [21,23,47]. Using low-coverage oligonucleotide arrays, the abundance of Ty elements has been estimated in different S. cerevisiae strains relative to that in the S288c reference genome [48]. The authors could not, however, distinguish between the diverse Ty families (Ty1, Ty2, Ty4 and Ty5 copia-like elements and gypsy-like Ty3), although this problem can be overcome by hybridizing Affymetrix yeast tiling arrays that provide complete and redundant coverage of the ∼12 Mb S. cerevisiae genome ([49] and Schacherer J., personal communication).
The fraction of Ty elements could be estimated from low-coverage sequencing of S. cerevisiae and S. paradoxus strains (genome resequencing project, [50]), but whole genome sequencing of S. cerevisiae strains RM11-1a (http://www.broad.mit.edu/annotation/fgi), YJM789 [51], AWRI1631 [52] and EC1118 [53] has also contributed to a better knowledge of Ty-related diversity and of insertion site polymorphism. However, due to the aforementioned difficulties produced by TE in genome assembling, some of these data still remain preliminary [51,52]. As concerns TE insertion polymorphism between individuals, rapid and extensive results are expected from the recently developed global TE mapping methods, based either on selective isolation or enrichment for TE-related loci followed by their physical mapping using microarrays [54,55] or combined with sequencing [56]. Data from the application of these approaches to large-scale investigations are now available for the polymorphism of human L1 LINE insertion sites in individuals [57,58], but are not yet available for yeast intraspecific studies.
Work on TE-related intraspecific variability has not been restricted to descriptive investigation: for instance, the insertion site polymorphism between strains is a good indication of the potential activity of the corresponding TEs [21,23], except for TE copies mobilized by homologous recombination as repeated sequences or being randomly lost. Investigation of the TE content of strains and isolates contributed to uncovering significant aspects of TE evolution. An example is the detection of Ty2-like elements in some S. mikatae strains highly similar to those in S. cerevisiae, therefore suggesting transfer from one species to the other [45]. Its generalized presence in S. cerevisiae strains indicates either an ancient introduction or a greater success (higher transposition rates and/or fewer element losses) than in S. mikatae. Analyses of the variability in TE contents between strains and isolates has also contributed to our understanding of the complexity of Saccharomyces diversity both in its natural environment (for example Ty distribution seems to correlate with the geographical origins of S. paradoxus isolates [45]) and in industrial strains. Indeed, a large proportion of industrial strains are hybrids [59]. According to their Ty content, strains of the hybrid species S. pastorianus could be classified into two groups [45] which turned out to correlate with two distinct interspecies hybridization events as suggested in [60]. Such studies also provide strain signatures: wine strains are characterized by lower Ty contents than S288c-related lab strains and pathogenic strains [39,61] and by inversion of the Ty1/Ty2 ratio with respect to the reference strain S288c [53,62].
Knowledge about the TE landscape at the intraspecific level is of great importance to our understanding of the impact of TEs on genomes. Before addressing this important issue, note that early studies were mainly focused on the presence of full-length TEs and did not consider their precise nucleotide sequence, which is useful to predict their putative autonomy. Therefore, the landscape of TE relics (especially solo-LTR) and the ratio of autonomous/nonautonomous TEs have been only poorly explored. However, data of this type may help elucidate several aspects of the relationships between TEs and their hosts: their effects on: (i) host genome stability, either as repeated sequences or by inducing local epigenetic changes; (ii) the host transcriptome, in addition to the direct effects mediated on adjacent sequence expression by the transcriptional regulatory motifs carried by TEs; and (iii) the dynamics of TE mobility and persistence [63].
4 Impact of yeast Transposable elements on their hosts: genome instabilities and altered transcriptomes
Karyotype changes due to chromosomal rearrangements are an additional source of inter- and intraspecific variability, and–not surprisingly–repetitive TEs have been found to be associated with a substantial proportion of these chromosomal rearrangements (see [64] for an historical example in S. cerevisiae and [65] for an extensive study of spontaneous mutations). LTR retrotransposons, the most abundant yeast TEs, may particularly threaten the integrity of the genomes they inhabit, as they are involved in the ‘mobilization’ of genome fragments in several ways. Homologous recombination between Ty-related repeats (and the often associated tRNA genes) has been well documented and the mechanisms and consequences of ectopic recombination resulting in polymorphism at Ty insertion sites (presence of a full-length Ty element or a solo-LTR) or in chromosomal deletion, duplication, inversion and translocation have been reviewed in [66]. The most recent findings concerning how TE-related recombination has shaped yeast genomes includes the results of investigations of interspecies hybrids, and in particular the lager-brewing yeast S. pastorianus and the wine yeast strains derived from S. cerevisiae–S. kudriavzevii crossings. These studies suggest that recombination between Ty sequences was involved in building their chimerical chromosomes [60,67]: some of the rearrangement breakpoints coincide with the presence of Ty-related sequences in the reference strain S288c. However, these reports did not confirm that Ty-related sequences are indeed present at these genomic loci in the corresponding parental and hybrid strains, and the whole-genome sequence of another S. pastorianus strain shows orthologous genes rather than Ty-sequences at the breakpoints [68]. Consequently, further investigations of this issue are required. In L. kluyveri and L. thermotolerans, some Ty-relics have been found in the vicinity of genes that obviously originate from horizontal gene transfer [69]. This suggests that the presence of TEs may facilitate the integration of foreign DNA, possibly because of the high local instability they generate.
Interestingly, several studies report that Ty-related chromosomal rearrangements (especially segmental duplications and gene amplifications) increase the fitness of strains under strong selective pressure, for example lab-evolved strains [70–72] and natural flor strains [73]. This suggests that genomic Ty-clusters are not distributed randomly but make up, in some cases, “particular genomic architecture [that] has been retained to facilitate evolvability of S. cerevisiae populations” [72]. Some of these rearrangements are reversible [72], indicating that TE-mediated adaptation can be particularly flexible.
Reverse-transcription of cellular mRNA is another mechanism for mobilizing genomic sequences: Ty-related reverse-transcriptase activity promotes retroposition and with homologous recombination, this can result in processed pseudogenes or functional retrogenes [74,75], and cDNA-mediated chromosomal rearrangements [76].
TE-related polymorphisms (insertions and rearrangements) constitute one of the genetic bases for phenotypic trait variations in yeasts. First, as mutagens, TE insertions can interrupt a gene in a particular strain and this can have pleiotropic effects, as exemplified by the insertion of a Ty1 element at the end of the HAP1 transcription factor in strain S288c. This single polymorphism results in unique expression patterns, different to those in other non-S288c lab strains and industrial strains [77], although this may not explain all the unusual phenotypic characteristics of this strain [78]. TEs and in particular Ty-elements can act as gene promoters [79,80]. As with mammalian TEs [81], Ty- and Tf-LTRs carry transcription start sites [82,83] suggesting that retrotransposon transcription has a significant influence upon the transcriptional output of yeast genomes. They also carry regulatory signals that modify adjacent gene expression and regulation [82,84–87].
Altogether, these studies suggest that TEs in yeast genomes may be selected for generating genomic variability in response to environmental stimuli, as stress conditions are known to favor TE expression [86,87], transposition and TE-mediated rearrangements [66]. Therefore, yeast TEs contribute to genome evolution, not as a driving force for speciation [88,89] but through genetic innovations contributing to strain fitness during colonization of new ecological niches. This can, in turn, result in geographical isolation.
5 Host–Transposable elements interactions and Transposable elements persistence in yeast genomes
TE invasions are initially harmful for the hosting organisms, and therefore TEs present in modern genomes are believed to have reached a steady state of equilibrium: their expansion, and ectopic recombination between TE copies, are limited by coevolution of TE traits and host regulatory mechanisms. A well-described TE trait that limits harmfulness is integration site specificity and one mechanism for such specificity has been illustrated for Ty5 insertion in heterochromatin [90]. A highly developed form of host–TE co-evolution consists in TE exaptation. This is an evolutionary process in which a characteristic that evolved under natural selection for a particular function comes under selection for a different function. Two examples have recently been reported in yeasts: (i) in K. lactis, a yeast species that lacks the HO endonuclease gene, the Alpha3 gene was demonstrated to be a transposon, the transposase activity of which is required to initiate formation of the DNA double-strand break facilitating mating-type switching [91]; (ii) In S. pombe, the transposase-derived CENP-B proteins are involved in host genome surveillance and mediate Tf elements silencing by sequestration of the corresponding genomic regions into heterochromatin through histone modification [92].
A remarkable diversity of genes and regulatory mechanisms dedicated either to repressing Ty element transposition [93–95] or to suppressing retrotransposon-mediated instability have been described [94,96]. In eukaryotes, RNAi is a major component of the defense against nucleic acid invaders such as viruses and TEs [97,98]. RNAi has been described to repress Tf2 expression moderately in S. pombe [99] and to decrease retrotransposon transcript abundance and mobility in C. neoformans [100]. Until recently, Saccharomycotina yeasts were believed not to have RNAi. However, siRNA have now been discovered in some budding yeast along with a noncanonical RNAi pathway involving an unusual Dicer, which particularly targets Ty elements [101]. No such an RNAi pathway is present in the model yeast S. cerevisiae, raising the issue of how Ty and Ty-host coevolution has been influenced by the loss of this ancient defense mechanism. This issue could be addressed by deep investigations of retrotransposon contents within species either having or lacking RNAi pathways. As concerns TE–host interactions, it is of particular interest that recent studies on transposition mechanisms have revealed the potential dual role of XRN1. It enhances Ty1 transposition through degradation of an interfering cryptic unstable transcript [102] and may be involved in VLP assembly in association with other RNA decay components [103–105]. This raises questions about the dynamics of Ty RNA storage/degradation, in view of its dual role as template for translation and reverse-transcription, and how Ty RNAs addressed to storage/encapsidation sites are distinguished from cellular RNAs addressed to degradation.
6 Concluding remarks
This review of recent work shows a strong bias of TE content in Saccharomycotina and especially of the Ty retrotransposons. Therefore, it is unlikely that the full diversity of yeast TEs has yet been revealed and fascinating results are expected from ongoing genome investigations of Taphrinomycotina, Ustilaginomycotina, Pucciniomycotina and Agaricomycotina yeast lineages and from the intraspecific investigations that have became feasible because of NGS technologies. The genetically tractable yeast S. cerevisiae will undoubtedly continue to be central to progress in understanding the associated mechanisms of host–TE interactions. Indeed, the heterologous RNAi pathway from S. castellii has been successfully (re)introduced into S. cerevisiae [101] and it can be used for transposition of heterologous TEs like LINE Zorro3 [106].
Disclosure of interest
The author declare that he has no conflicts of interest concerning this article.
Acknowledgements
We would like to thank all our colleagues from the Génolevures consortium for helpful discussions. This work was supported by funding from CNRS (GDR 2354, the Génolevures Consortium) and ANR (ANR-05-BLAN-0331, GENARISE).


