1 Introduction
The phenotype 〚URE3〛 in the yeast Saccharomyces cerevisiae 〚1〛 is due to the protein Ure2 that is encoded by the chromosomal gene URE2 〚2〛. This phenotype corresponds to a dominant and heritable loss of the function of Ure2p. The 〚URE3〛 phenotype is inherited in a non-Mendelian fashion 〚3〛. It is dominant, stable, transmissible by cytoduction and reversible by treatment of yeast cells by chemicals, such as guanidinium chloride, that interact with proteins.
Ure2p has a molecular mass of 40.4 kDa and is made of 354 amino acid residues. Ure2p and PrP are unrelated in primary sequence. They are however both at the origin of dominant heritable changes. The function of Ure2p is unknown. All we know about its function is that it is involved in a signal transduction pathway that regulates nitrogen catabolism in yeast cells 〚4〛.
Our group and others demonstrated previously that Ure2p is composed of two distinct regions 〚5, 6〛. The N-terminal region is poorly structured and sensitive to proteolysis, while the C-terminal region of the protein is compact and relatively resistant to proteolysis 〚5〛. We defined the physical boundaries of the two regions. The N-terminal region extends from amino acid residues 1 to 93 〚5〛. This region has a peculiar amino acid composition. It is rich in asparagine and glutamine residues (43 of 93 amino acid residues) and in serine and threonine residues (13 of the remaining 50 amino acid residues). It is sufficient to induce de novo generation of the 〚URE3〛 phenotype 〚7, 8〛. The C-terminal region extends from amino acid residues 94 to 354 〚5, 6〛.
2 Results
Ure2p is a soluble and dimeric protein 〚5, 6〛. Soluble Ure2p forms high molecular weight oligomers that are visible in the electron microscope 〚5〛. The diameter of these particles is about 12 nm (figure 1A). Ure2p assembles in vitro into insoluble fibrils that are on average 15 nm wide and over 1 μm in length 〚5〛 (figure 1B). Assembly of Ure2p into fibrils can be monitored by a variety of spectroscopic methods among which is Congo Red binding 〚5, 9, 10〛. Congo Red binding to these fibrils is accompanied by a green birefringence of polarized light. These fibrils are therefore amyloid fibrils. The limiting step in the assembly process of Ure2p into amyloid fibrils is the nucleation step. Indeed, when preformed fibrils are sheared and diluted 10 000-fold into a solution of soluble Ure2p, the lag phase preceding Ure2p assembly, during which nuclei form, disappears 〚5〛 (figure 1C).

Soluble Ure2p assembles into high molecular weight oligomers. A, Soluble ring-shaped Ure2p oligomers. B, Insoluble Ure2p fibrils. The bar in panel A and B represents 100 nm. The bar in the inset in panel A corresponds to 20 nm. C, () time course of Ure2p (10 μM) assembly into fibrils monitored by Congo Red binding; (■) assembly of Ure2p (10 μM) into fibrils upon addition of preformed fibrils (10 nm). Adapted from 〚5〛.
Assembly of Ure2p into amyloid fibrils is accompanied by an increased resistance to proteolysis 〚5, 9〛. Indeed, the soluble form of Ure2p (0.8 mg/mL) is degraded within one minute by proteinase K (2.4 μg/mL at 30 °C) while the assembled form of the protein resists the same treatment for over 8 min (figure 2).
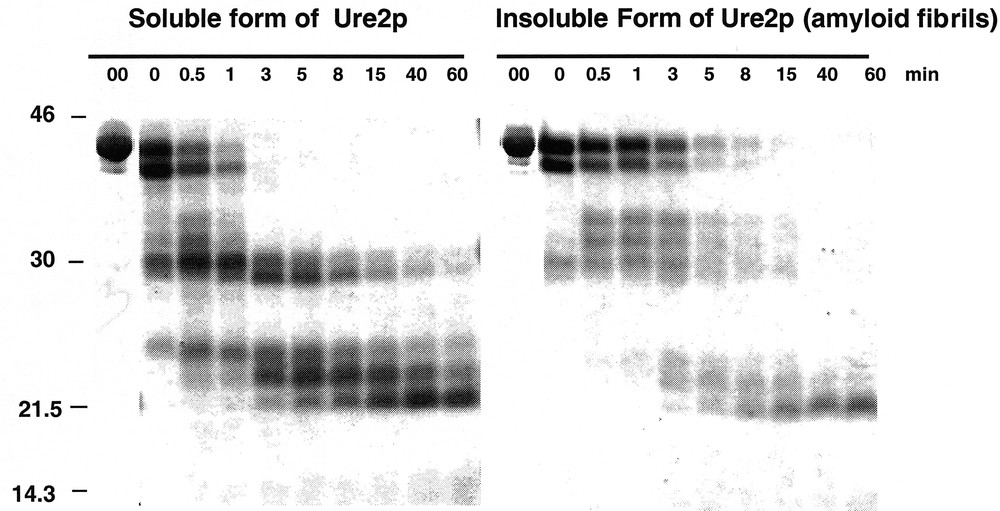
Assembly of Ure2p into amyloid fibrils is accompanied by an increased resistance to proteolysis. Time courses of degradation of the soluble (left panel) and insoluble (right panel) forms of Ure2p (0.8 mg/mL) by proteinase K (2.4 μg/mL) monitored by SDS-PAGE. Adapted from 〚5〛.
The α-helical content of Ure2p and that of its C-terminal region are identical 〚10〛. The spectra are perfectly superimposable between 250 and 210 nm. The CD spectra differ significantly between 210 and 195 nm. The circular dichroism difference spectrum of Ure2p and its C-terminal region indicate a large decrease of the ellipticity signal between 205 and 195 nm. This observation indicates that the N-terminal region of Ure2p is poorly structured 〚10〛 (figure 3).

Ure2p N-terminal region is poorly structured. The circular dichroism difference spectrum of full length Ure2p and Ure2p C-terminal region reveals a decrease of the ellipticity signal between 205 and 195 nm. Adapted from 〚10〛.
The C-terminal region of Ure2p is dimeric in solution as is full length Ure2p. This indicates that Ure2p dimerizes through its C-terminal region 〚5, 6〛. The C-terminal region of Ure2p has the same fold as full length Ure2p. Indeed, the degradation patterns of full length Ure2p and its C-terminal region by proteinase K are identical 〚10〛 (figure 4). In addition, the two polypeptides show superimposable equilibrium unfolding curves which indicates not only that they have the same fold but also that the N-terminal region of Ure2p does not increase the overall stability of the protein 〚10〛. Ure2p unfolding is a two-step process in a range of pH conditions 〚10〛 (figure 5). The unfolding intermediate revealed by the guanidinium chloride equilibrium unfolding studies is stable and dimeric 〚10〛. This unfolding intermediate does not form amyloids 〚10〛. It is therefore not a precursor of amyloid fibrils. Another folding intermediate obtained in a range of pH conditions is a precursor of amyloid fibrils. Indeed, amyloid fibrils form at pHs between 5.5 and 7.5, while unstructured aggregates form either at higher or lower pHs 〚10〛.
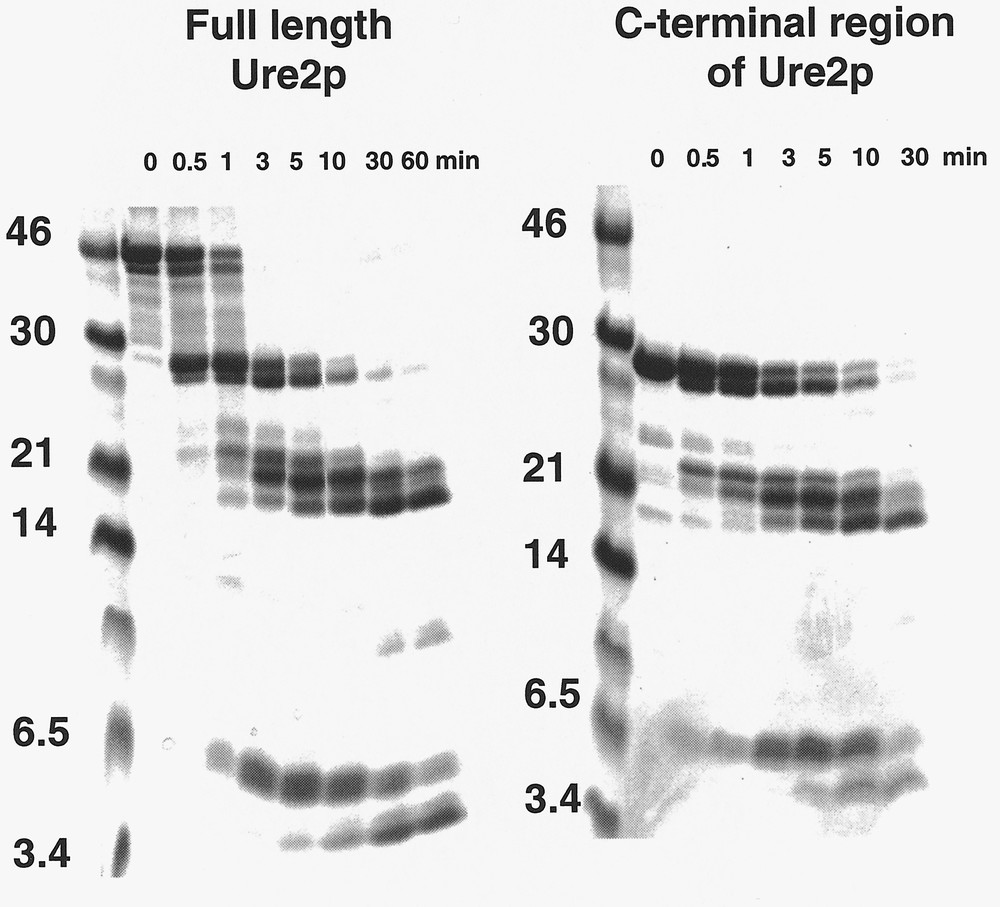
Time courses of degradation of full-length Ure2p and its C-terminal region by proteinase K. The degradation patterns are identical indicating that the two polypeptides have the same fold. Adapted from 〚10〛.
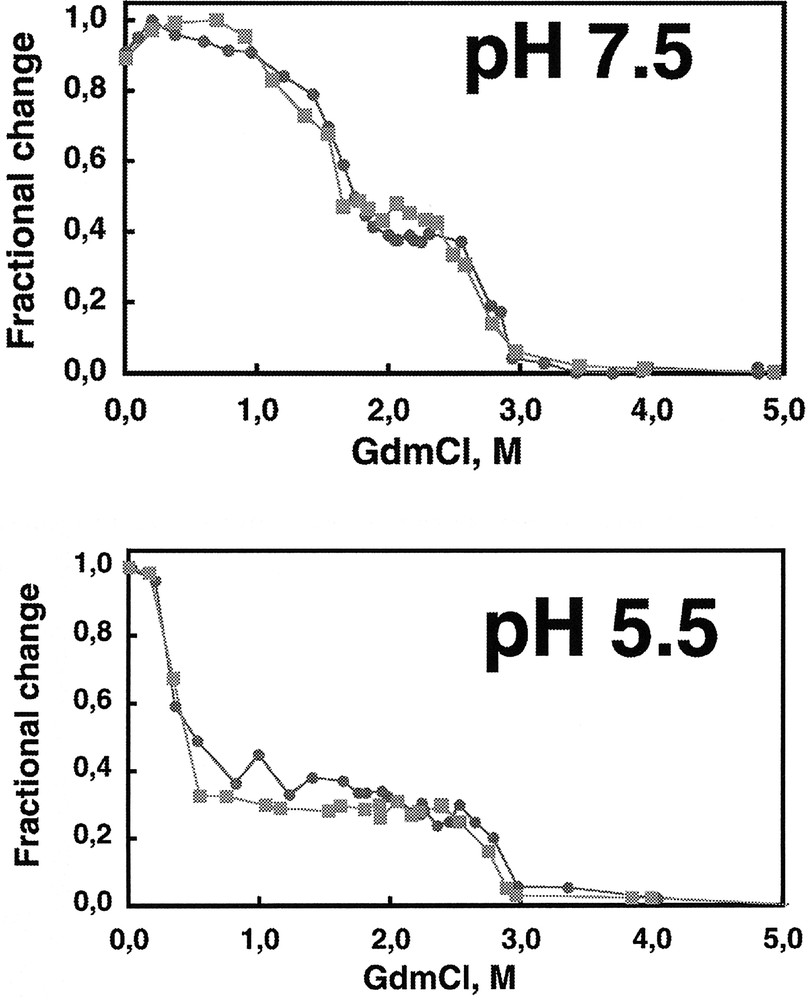
The equilibrium unfolding curves of full length Ure2p () and its C-terminal region (■) by guanidinium chloride at various pHs are superimposable and reveal the existence of an unfolding intermediate. The two polypeptides have therefore the same fold and the N-terminal region appears not to increase the overall stability of the protein. Adapted from 〚10〛.
To better understand the molecular events that generate an amyloid fibril assembly competent form of Ure2p, it is crucial to determine the three dimensional structure of the protein in its soluble and assembled forms.
We have solved the crystal structure of the structured part of Ure2p i.e. the C-terminal region of the protein 〚11〛. This is the first crystal structure of a protein with prion properties. The dimeric state of Ure2p is confirmed by the crystal structure 〚11〛 (figure 6A). The monomers colored in blue and green are head to tail. The polypeptide chain comprises two domains separated by a short linker region in yellow. The N-terminal domain is made of a four-stranded β-sheet flanked on each side by two α-helices. The C-terminal domain is all α-helical (figure 6B).

Ribbon representation of the three dimensional structure of the globular region of Ure2p in its dimeric (left) and monomeric (right) forms. Adapted from 〚11〛.
The C-terminal region of Ure2p shares a very low degree of sequence similarity with a class of enzymes involved in detoxification, the glutathione S-transferases 〚2〛. A structural comparison of the two molecules reveals they have nevertheless a similar fold 〚11〛 (figure 7A). Ure2p differs significantly from GSTs by its 110 N-terminal amino acid residues and a 32 amino acid residues insertion we called α-cap that is highly flexible. Indeed, the carbon backbone of Ure2p monomers within a dimer or in the different crystal forms we obtained superimpose perfectly apart from the cap region 〚11〛. This indicates that the cap region adopts different conformations in Ure2p monomer within the dimer and in the different crystal forms we obtained. The difference measured at amino acid residues 284–286 indicate that the cap region moves by up to 6.2 Å which corresponds to a rotation of about 25° 〚11〛 (figure 7B).
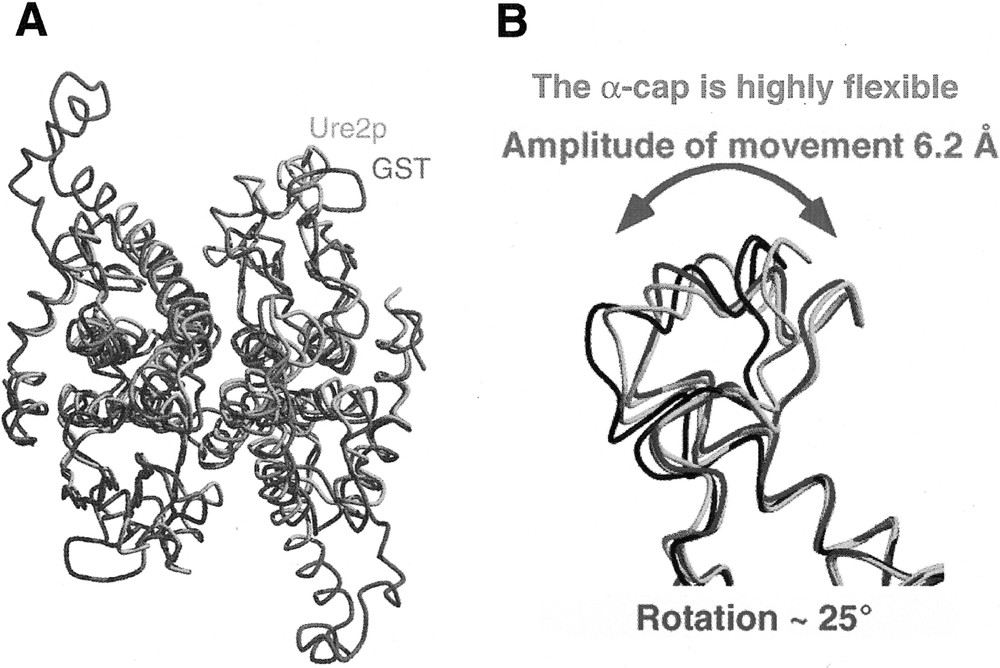
A, Structural comparison of Ure2p (blue and orange) and bacterial GST (purple). B, Structural comparison of the Ure2p cap region in different crystal forms of the protein. Adapted from 〚11〛.
The structure we have solved reveals that the 90 N-terminal amino acid residues of one Ure2p monomer are probably located in the vicinity of the cap region of the partner monomer in the dimeric form of the protein 〚11〛. This suggests that the two polypeptides could interact transiently. Such an interaction could modulate the assembly properties of Ure2p.
3 Conclusion
As in the case for PrPc whose structure was solved nearly five years ago by Wüthrich and coworkers 〚12–13〛, the three-dimensional structure of Ure2p opens the way for the understanding of the molecular events that lead to the invasive behavior of prion proteins. However, a crucial piece of data is still lacking to generate a full picture of the molecular events at the origin of prion propagation. Indeed, as for PrPc, the structure of the flexible and/or poorly structured N-terminal region of Ure2p is not yet determined. This region is required for the propagation of the 〚URE3〛 phenotype, i.e. the transmissible loss of the function of Ure2p. Thus it is crucial to determine one or a subset of conformations of this region in solution either in protein cystals or by NMR.
A number of other issues need to be addressed such as the role of Ure2p dimerization and that of the α-cap in amyloid formation. It is also crucial to determine the structure of Ure2p in amyloid fibrils in order to understand the conformational changes at the origin of amyloid formation.
Acknowledgements
Supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer and the French Ministry of Research and Technology. We thank Prof. Joël Janin for helpful discussions.


