1 Introduction
In biological databases, data on gene function is still driven by the hypothesis of Beadle and Tatum “one gene = one enzyme” or the revised model “one gene = one peptide”, which proposes that a gene is the entire nucleotide sequence required for the synthesis of a functional polypeptide or RNA 〚1〛. For example in the OMIM database (http://www3.ncbi.nlm.nih.gov/Omim/), the three GAD genes 〚GAD1(OMIM entry: 605363), GAD2 (OMIM entry: 138275 ) and GAD3 (OMIM entry: 138276)〛 are all identified as glutamic acid decarboxylase (EC number: 4.1.1.15). Nevertheless, although they share extensive similarities, GAD1 and GAD2 are respectively involved in the autoimmune disease stiff man syndrome and in insulin-dependent diabetes mellitus, which are clinically distinct; a seven amino acid difference between the isoforms is believed to account for these differences. This illustrates the limited usefulness of the “one gene = one enzyme” model to account for functional attributes. In addition, assigning a role to an individual gene that encodes one particular subunit of a multimeric complex could be misleading, even within the framework of the revised “one gene = one peptide” model. For example, guanine nucleotide-binding proteins are heterotrimers that mediate the release of hormones. Five functional classes exist, which are either stimulatory (Gs) (OMIM entry:139320) or inhibitory (Gi) (OMIM entry: 139310) GTP-binding regulators of adenylate cyclase, phototransductors in retinal rods (transducin 1) (OMIM entry: 139330) and cones (transducin 2) (OMIM entry: 139340), and a class of unknown function, which is abundant in the brain (Go) (OMIM entry:139311). Each of them has a unique α chain, but the β and γ chains appear to be identical, which suggests that functional specificity is only due to the α chain gene. Thus, the function of the β and γ peptides cannot be implemented individually, without considering the heterotrimeric unit they compose. The inadequacy of the “one gene = one peptide” model is obvious given the alternative splice patterns (3’ or internal exons) and the post-transcriptional and post-translational modifications, which result in products with a variety of structures and/or functions. Accordingly, alternative splicing means that the BCLX (OMIM entry: 600039) gene positively and negatively regulates programmed-cell death: the larger splice product (BCLXL) inhibits cell death in the absence of growth factor, whereas the smaller one (BCLXS) counteracts the ability of BCL2 to enhance cell survival. Numerous examples emphasize the complexity of ‘gene physiology’ (i.e. gene function), which carries out and coordinates biological functions by means of genetic and epigenetic regulation pathways 〚2〛.
To overcome the limits of the classical gene model, which refers to static structures, an expanded constructionist gene concept has been proposed to account for the function of a particular DNA sequence in a developmental system 〚3〛. This concept was renamed the ‘molecular process gene concept’ to facilitate the replacement of the classic molecular gene concept. The molecular process gene concept does not just identify a gene based on its DNA sequence alone. Instead, it also studies the role of this DNA sequence. This idea is similar to Goldschmidt’s one, who rejected the static concept by stressing the role of the environment on gene physiology 〚4〛. These views shifted the emphasis from genetic structures to developmental processes. They also lead us to reconsider the simplistic reductionist theory, which claimed that function was based on structure and that there is a one-to-one relationship between structure and function. In this respect, gene function modelling is more limited by the problem of function modelling (i.e., how to represent self-organising components that develop and change within their bearer system) than gene modelling. In other words, if we do not distinguish between gene function modelling and gene modelling, regardless of the gene concept 〚5〛, this will elude the central debate concerning the requirement for an alternative paradigm, as the reductionist one is ill-adapted to functional representations. An example of this dilemma is provided by the delineation of the molecular process gene concept, which is in fact a shift from the molecular gene concept to a dynamic framework (dealing with a succession of facts to pass from one state to another) rather than a new concept in the reductionist paradigm (concerned with established systems).
Living systems are continuously being transformed from one state to another. Thus, there is a permanent problem of how to achieve and to coordinate biological functions so as to maintain homeostasis or to lead to differentiation, under the constraints of exogenous and/or endogenous fluctuations. Without making any assumptions concerning the mechanisms (physical laws) involved in the maintenance of these organisations, biological functions aim to maximise survival and reproduction at levels in the biological hierarchy. This introduces the teleological concept, which is important for function modelling. This concept, in which the goal is to accomplish a particular biological function, has been the source of much debate since Aristotle’s time. The major point for controversy is whether the goal is exogenous (described as being a metaphysic being endowed with a purpose by Aristotle and his followers) or endogenous (the concept of strategy within the systemic paradigm).
This controversy between the teleological and non-teleological meanings of function stems from the improper bringing together of two distinct concepts of function. The etiological concept aims to explain the acquisition of a function via natural selection 〚6〛; it is quite distinct from explaining the function itself by showing (i) how function is instantiated in the entities that have it and (ii) how its instantiations are articulated to achieve a particular end 〚7〛. By incorporating these two approaches, the concept of function is no longer an ontological property. Instead it is a relational property that controls all abiological and biological factors in a particular environment. This in turn gives rise to emergent properties – functions – that appear in the form of finalised processes 〚8〛. Nevertheless, this relational nature of the function goes against some of the fundamental concepts of the current paradigms within the notion of biological functions. Most notably, one of the main weaknesses of the analytical paradigm (i.e. the reductionist or the classical mechanics paradigm) is that they are often unaware of the Aristotle’s principle: “The whole is more than the sum of the parts”, whereas biological functions result from the emergent properties of the self-organised components. For example, ribosomal subunits can be reconstituted from dissociated molecular components and are fully functional thanks to the emergent properties of the reorganised components 〚9〛; however, no theories can explain this observation in the analytical paradigm based on the invariance of the structure 〚10〛. Although the thermodynamic paradigm and the Darwinian paradigm of evolution are based on the assumption of the morphogenesis of the structure over time, they deal with the morpho-dynamics of the structure rather than with their functional kinematics. This is undoubtedly the reason why they have rarely been in functional modelling. To overcome this problem, the structuralist paradigm was derived from a common interest for the kinetic structure-function and the dynamical structure–evolution relationships. However, subsequent developments quickly revealed that this model had internal limitations due to the closure constraints required to make the description exhaustive (according to the fourth Cartesian precept of the Discours de la Méthode). In this respect, it is questionable how relevance the structuralist paradigm is for the modelling of functions. This was shown by Piaget in 1968 who proposed (at the same time as Monod introduced the concept of microscopic cybernetics in biology 〚11〛) to broaden the structuralist paradigm to the cybernetic one. The methodological effectiveness of the cybernetic paradigm was based on its two key concepts: behavioural blackbox and teleological feedback. Instead of wondering about the internal composition (“Of what is it made?”), the emphasis was on the functioning and the functions of the system in a known context (“What it does, in what, why?”). But, like structuralism, the cybernetic paradigm quickly reached its operational limitations due to the closure properties of the context-free, structural and functional stability of the blackbox and of the stability of the system.
In this article, we present the conceptual framework of the systemic paradigm, its formalism and its relevance to the modelling of biological functions. In addition, we provide some examples and current perspectives on the modelling of biological facts to drive data acquisition.
2 The systemic paradigm and its formalism
“A paradigm contains, for any speech being carried out under its empire, the fundamental concepts or the main categories of intelligibility, as well as the type of logical attraction/repulsion relations (i.e. conjunction, disjunction, implication, etc.) between these concepts and categories” 〚12〛.
The basic concept of the systemic paradigm is not the object or the combination of stable objects (i.e. the structure) but the action 〚13〛. The systemic paradigm aims to answer the following questions: what does it do? what are the functions, transformations and operations done or that need to be done? Conversely, the analytical paradigm asks: what is it made of? what are the components, the objects or the organs that are combined to constitute the phenomenon?
The axiomatic of the conjunctive logic ensures the instrumentation of the systemic paradigm and helps to represent complex, non-decomposable systems. The three major principles are recursivity, synchronism (from Greek sun, ‘with’ and khronos, ‘time’) and diachronism (from Greek dia, ‘through’ and khronos). In the conjunctive logic, a system is not described per se, but in relation to a particular goal (recursivity); it is represented in relation to its external environment, according to its behaviour (operational teleology or synchronism); it is not perceived according to the totality of its components, but only according to the groups of active components that are functional to the ends sought (irreversible teleology or diachronism) 〚14〛. In contrast, the disjunctive logic leads to the decomposition of the system, so that its components can be analysed one by one.
The general system concept emerged through the conjunction of the two paradigms: (i) the cybernetic paradigm founded on the concepts of an active environment and of teleology, characterised by the general blackbox and feedback concepts, and (ii) the structuralist paradigm founded on the combination of the concepts of functioning and transformation. We present below the founding concepts of the systemic paradigm with its canonical representation in the form of a general system and the derived formalism. This general system concept was introduced by Bertalanffy to generalise the concept of an ‘open system’ and to overcome the limitations imposed by the mechanicist paradigm to the modelling of living systems 〚15〛.
2.1 The canonical form of the general system
The systemic paradigm is expressed correctly by an ideogram named the ‘general system’ because its description identifies the essential articulations of the reasoning: an object behaving teleologically in an environment, which carries on an activity and the internal structure of which changes with time, although its identity is preserved. This definition, involving five common concepts (see below), can be shared because it is sufficiently formalised. The system behaviour is represented by two inseparable components: the function (what is done) and the evolution (how to do it). The model of the form, which changes and ensures new functions, characterises the structuralist conjunction. The axiomatic of systemic conjunction proposes to hold inseparable the function and the evolution of one component or of a group of components in an active environment, with respect to the goals to which they are identifiable. This inseparability of founding concepts results in the conceptualisation of the general system as the representation of active components identified by an endogenous goal-directedness and an active environment in which they function and transform themselves (Fig. 1). In a more mnemonic way, the general system is described by an action (or several actions) in an environment (spatio-temporal context). This system functions and transforms itself as well as the environment. This infers that the canonical form of the general system is also the definition of a general system.
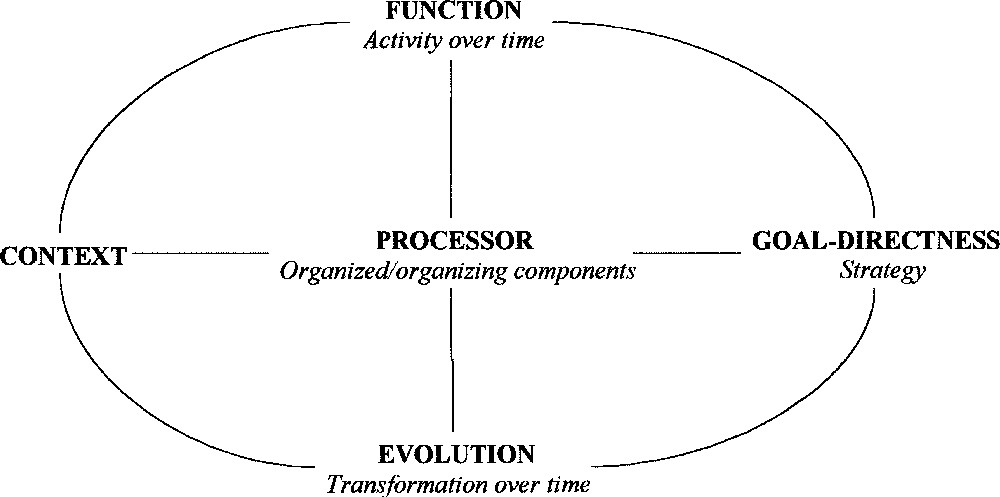
The canonic form of the general system. The systemic paradigm is the conjunction of the cybernetic (horizontal) concepts and the structural (vertical) ones. The lines represent the relationships between the concepts.
2.2 The systemic formalism
2.2.1 The canonical model of process
An action or a function can be characterised recursively; it conveniently fits the general process concept. A process is defined by its exercise and its result. A process occurs when it is possible to follow how an object’s position changes over time in a reference frame ‘space–form’: the combination of temporal transfer (how an object moves in a particular space over time; for example: transport between subcellular compartments) and the modification of form (a morphological change; for example: a post-translational modification by phosphorylation) constitute a process; it can be recognised as a displacement in the time–space–form (TSF) reference frame (Fig. 2). Form is used to describe organised and organising entities (tangible or not) within a process. An unique entity will be described differently according to its relationships with the context (organised), including actions on the environment (organising) and to itself (self-organising).
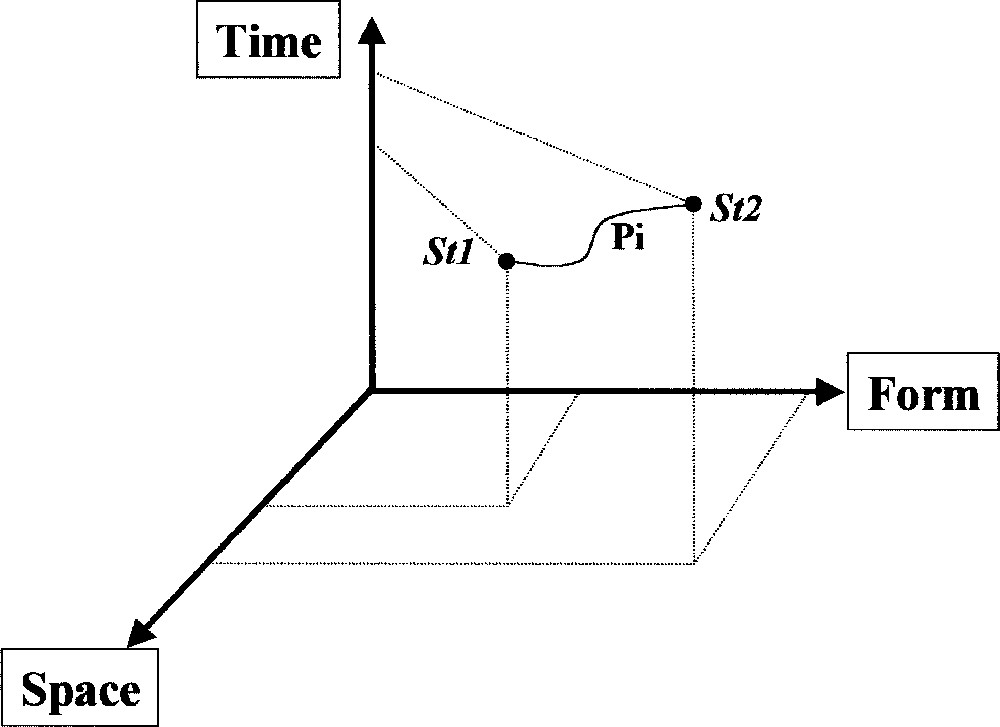
The canonical form of the process. A process Pr is applied to one entity that passes from state 1 (St1) to state 2 (St2) by irreversible changes in form (F) and/or location (S).
In this respect, a process is a multiple and tangled complex action, that is perceived as a change in the TSF reference frame. This made it possible to account for the articulation of the three prototypical functions: the function of temporal transfer (storage, memorisation, etc.), the function of morphological transformation (processing, computation, etc.) and the function of transfer in space (transport, transmission, etc.). These functions are exerted on tangible or intangible entities.
This allows us to introduce the concept of complex systems, which is essential to the description and the representation of the functioning of living systems: any complex system is represented by a system of multiple actions, by a process or by a tangle of processes. Even if these actions are very tangled, they can always be represented by the composition of temporal, space (transfer) and morphological (transformation) modifications.
2.2.2 Network and feedback: complex system and interrelationships of actions
Pr (processor) is used to indicate the blackbox in which a process occurs and each processor is characterised by the designations allotted to its input and its output. The interrelationships of n processors, identified by the functions that each one performs, will allow a reticular representation of the complex system. There is a relationship between two processors Pi and Pj, when one of the outputs of Pi is an input of Pj; in this case, the relationship (Pi, Pj) is activated. All the combinations of interrelationships between n processors can be represented by use of the structural matrix of the system: the presence of a ‘1’ at the intersection (Pi, Pj) means that the interrelationship is activated, the presence of a ‘0’ means that this interrelationship is prohibited. There are (2n)2 different ways of filling a square matrix with size n of ‘0’ and ‘1’. In other words, there are (2n)2 possible different interrelationships and (2n)2 different behaviours. This is also known as the variety 〚16〛 of the system. Feedback interrelationships can be predicted by active relations (‘1’) below the principal diagonal of the matrix; they express a feedback as some of the inputs of the processor of interest are the output of this same processor. These feedback relationships are essential, because they provide upstream processor information on the behaviour possibly induced by a downstream processor. The frame made of processors connected by interrelationships represents the network of the system.
A graph in which nodes are the processors and the directed edges are the interrelationships constitutes a traditional and very general system model. This representation makes most of the resources of graph theory available for systemic modelling, including with the concept of graph matrix introduced here under the term of structural matrix. As the number of processors is high, the structural matrix has a quasi-decomposable structure and appears to be made up of sub-matrices. This enables us to differentiate the system in as many subsystems or levels, each level being modelled by its network and being interpreted in a relatively autonomous way, if the coupling relations between sublevels were carefully identified.
3 Systemic modelling of biological functions
Biological functions are performed by complex systems organised in a hierarchical (molecular cellular, organismic, etc.) and inclusive (cellular including molecular) manner. The term ‘complex system’, refers to irreducible units that are actively organised (i. e., the action of organisation and the result of this action) and can be conveniently qualified by the processor of the systemic formalism.
To describe the intelligibility of the TGFβ-dependent signalling process 〚17〛, one has to deal with irreversible changes over time concerning form (activation) and localisation (subcellular compartment). These changes recursively depend on environmental changes, which in turn induce transformation within the organisation of signalling components. Molecular interactions qualified in the TSF triad as elementary processes account for a dynamic representation of the TGFβ signalling.
At the organismic level (for example, during embryonic development), TGFβ signalling is one of the components of a more complex unit, including VCC/β-catenin signalling, which carries out the dorsal–ventral polarisation of the embryo; accordingly, it can be globally modelled as a processor that cooperates with VCC/β-catenin signalling, another processor, to set up the dorsal–ventral axis process.
Thus, the description of the processors according to the systemic formalism makes it possible to establish an operational connection between the systemic paradigm and the modelling of the biological functions. With this intention, the design of elementary processors of the TGFβ signalling was considered according to the specificity related to biological systems.
3.1 The time problem: dynamic versus static modelling
The lack of awareness of the temporal characteristics of any functioning process in biology is quite dangerous, to quote Van Regenmortel 〚18〛: “Thinking in terms of static structures can affect the way we imagine the process of biological recognition to occur. Static images tend to reinforce the appeal of lock- and key-models for describing and explaining molecular recognition. 〚...〛 They also make it more difficult to conceive of recognition as a process. 〚...〛 By transforming a process or a relationship into a fixed thing (〚...〛 a timeless concept) that can be abstracted from the interactive system, reification leads us to view recognition as a static phenomenon rather than an activity.”
In fact, living systems change irreversibly over time and while functioning, regardless of the hierarchical level in question (from the biochemical level to the population level). Nevertheless, they are often presented as static established systems. If the example of the TFGβ-signalling is taken again, the current representations correspond to a kinematic representation with time-ordered stages of the signalling pathway. This is completely different from a dynamic view, which includes the facts (changes in form, spatial transfer etc.) that allow the system to pass from one state to another. Using the systemic formalism, the combination of time with changes in space and form provides information about the states of the system and its evolution in a dynamic representation. Conversely, a synchronic view represents the time-ordered stages of the pathway.
3.2 The form problem: a unique entity and its multifarious forms
According to the systemic formalism, form changes refer to the different activities of one processor related to context variations. In our case, form changes concern all the modifications occurring in a unique entity. In addition to the topological organisation of molecules, which confers distinct functional properties from those predicted at the literal and linear levels 〚19〛, the molecules may exist in active or inactive forms, monomeric or multimeric forms, as multiple isoforms due to post-transcriptional and post-translational modifications, etc. In our example, MADH3 exists in a monomeric inactive form and different heteromeric active forms; all these are highly dependent upon their environment and notably the molecular interactions in which they are involved.
In the TSF triad, the component behaves as a unique entity and its various forms are taken into account together with localisation and time. In data acquisition, a practical consequence would be to link its corresponding identifying symbol to ontologies (or even controlled vocabulary) concerning all known modifications (see below, § 3.5). In other words, instead of entering data concerning all MADH3 forms, only the unique identifier would be referred to and the types of transformation achieved would be mentioned (activated, complexed, phosphorylated, etc.).
3.3 The localisation problem: depending on the location, depending on the function
Subcellular localisation plays an important role in the activity of many compounds. It concerns both space, as predicted by the analytical model, the whole environment in which the compound is embedded and the successive forms taken by this compound. For example, when activated, MADH3 behaves as a signal transducer in the cytoplasm, whereas it behaves as a gene-specific transcriptional factor in the nucleus. In addition, the localisation specifies what part of the molecule will be active. The L3 loop of the C-carboxyl MH2 domain on the cytoplasmic membrane determines the specificity of the MADH3 interaction with the type I receptors, whereas its nuclear accumulation depends on a nuclear localisation-like sequence (NLS-like) in the N-terminal region. In addition, the N-terminal MH1 domain of MADH3 can bind to specific DNA sequences, termed Smad-binding element (SBE), to interact with proteins (reviewed in 〚20〛). These data emphasize how it would be misleading to assess function without considering localisation.
3.4 Network models: the reality is too complex to be represented in every details
The space of states that an entity (molecule, cell, etc.) can take is multidimensional and also depends on time, modified forms, subcellular location, developmental stages, etc. In other words, the processor for systemic modelling of biological functions is a vector with a certain combination of factors for all state dimensions (form changes, location changes, etc.). The network models consist of the processors represented at the nodes and the interrelations between processors at the directed edges of the graph. The network concept is an expansion of the pathway concept and recognises the presence of feedback, redundancy, cross talk, etc. There are three kinds of interrelationships linking these processors: (i) the linear causality relationships in which the causes precedes the effects and leads to them in a systematic way – generally a complex set of causes, which are usually independent, combines to produce one or several effects –; (ii) the retroactive relationships, which are characterised by a circularity between the processors; the anteriority of the cause in relation to the effect disappearing to give way to the regulator; (iii) the recursive relationships for which the produced effects are necessary to the processes which produce them. From an empirical point of view, the relationships of linear causality are rare, if they occur at all, as biological events are submitted to multiple feedback regulation loops. These feedback relationships can be positive and/or negative, depending of the ‘necessary behaviour’: negative regulations maintain things at or near the set point, whereas positive regulations moves things away from the set point; the negative regulation is kinematic and concerns established systems, the positive regulation is dynamic and corresponds to a succession of facts, with the transition from one state to another. In such a tangle of regulation loops, additional complexity is added to the system through the redundancy and the pleiotropy of networks to achieve a stationary state, which is associated with additional dynamic properties 〚2〛. Interrelationships of linear causality and feedback interrelations are the inputs and the outputs of processors; they delineate a particular ‘functional order’ to achieve a particular result.
Fifteen processors are presented as the simplified view of the role of MADH3 as a signal transduction component (Fig. 3). The processor P1 consists of the heteromeric complex made of TGFβ and TGFβ type II receptors, P2 represents the interaction between type I and type II receptors leading to the activation of the type I (*TGFBRI) through phosphorylation of Ser/Thr juxtamembrane sites; both complexes are at the cell membrane. The next step is the formation of a heterotrimeric complex (*TGFBRI:MADH3:SARA = P5) due to the presentation of MADH3 to activated *TGFBR1 by SARA (P4). These events are located at the internal membrane and within the cytoplasm. After becoming associated with MADH4, activated MADH3 (P6) is translocated to the nucleus, where it activates TGFβ-dependent genes by interacting with DNA-response elements (P11, P12, P13). These MADH3 activities may be inhibited by the Erk kinase in the cytoplasm (P3) and the proto-oncogene SNO in the nucleus (P14 and P15). Ubiquitin-mediated degradation of MADH3 plays a key regulatory role by switching off this inhibition (P8). Such modelling reveals a comprehensive pattern for the function of interest.
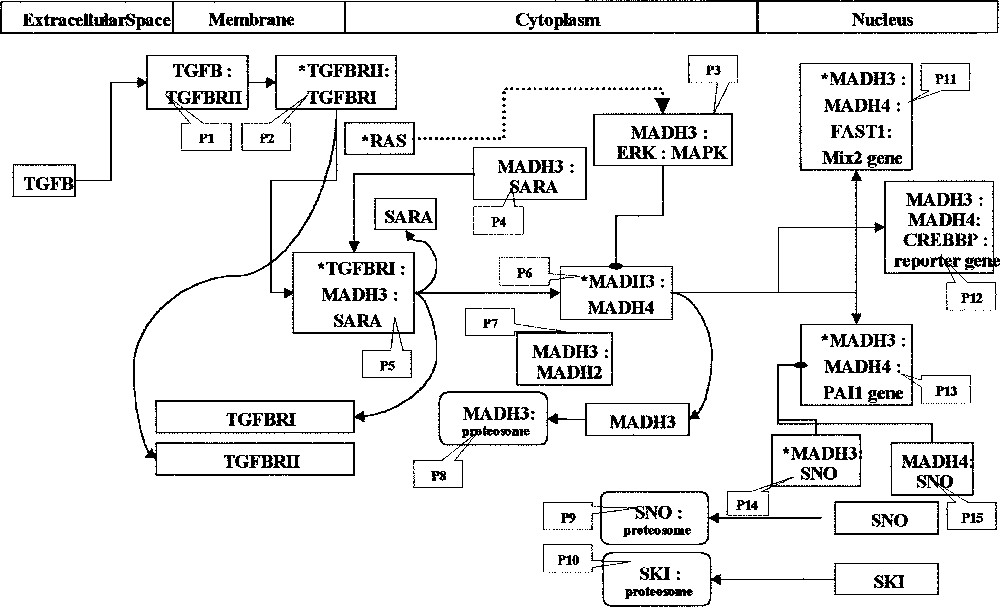
Interrelationships in TGFß-dependent signalling. The processors (open boxes) represent the molecular interactions; their composition is indicated by the corresponding gene symbol and stars indicate form changes; processors are numbered with grey boxes. The subcellular compartments are indicated at the top of the figure (see text for details).
Living systems are so complicated that we cannot represent every detail. Consequently, it seems more reasonable to work on maps, which show prototypical patterns of functions inferred from the grouping of selected structures and relationships on a functional basis (for example, the prototypical patterns of chromatin remodelling by the histone acetyltransferase/deacetylase group 〚21〛) instead of trying to be exhaustive, which is impossible. According to the systemic formalism, this approach aims to model the prototypes of the functions of interest and to build the corresponding sub-matrices connected through the adequate feedback relationships. This kind of strategy should drive data acquisition in comparative and functional genomics.
3.5 Example: a function-associated controlled vocabulary built with reference to the systemic formalism
The systemic formalism should be of great help to engineer our knowledge as its intelligibility is advantageous because it clearly refers to a given model. In the absence of this strong theoretical frame, the criteria for sharable knowledge remain poorly defined.
In this respect, one potential application is the delineation of a function-associated controlled vocabulary. The current biological databases use very heterogeneous function-associated vocabulary. This is one of the consequences of differences in the conceptual schemes of databases and the formats in which the data are presented, for example, if we consider the way in which the function of MADH3 transduction signalling is expressed in the databases. In Genbank (http://www.ncbi.nlm.nih.gov/Genbank/), only one of the ten entries (U76622) mentions that MADH3 is a mediator of TGF-beta family signal. In TrEMBL (http://www.expasy.ch/sprot/), which is the reference database for proteins, there is no entry yet for the human gene product and the entry for the mouse one does not mention any function. The use of natural language makes it difficult to exploit the data in the OMIM database (http://www.ncbi.nlm.nih.gov/; entry 603109). Finally, GenAtlas (http://www.citi2.fr/GENATLAS/; entry 19599) presents distinct complementary information. The specialised databases dedicated to signal transduction, Transpath (http://transpath.gbf.de/; entry M0000002332) and CSNDB (http://geo.nihs.go.jp/csndb/; entry M1270), display data on interactions and pathways, but it is not possible to determine the basic function of the well-characterised MADH3 gene product.
These examples shows the difficulty of finding the functional role of any entity in the absence of a theoretical framework to assert a representation and its sharing. Thanks to the systemic formalism, every processor can be described in the TSF triad by use of a constrained vocabulary. This vocabulary conveniently organizes our current knowledge into two main sections: (i) one dedicated to the physical entities (structures) divided into molecular structures and supra-molecular structures (including tissue-specific structures and sub-cellular components) and (ii) one dedicated to the activity information (processes) 〚22〛; these sections can be filled-in with files developed by the Gene Ontology Project 〚23〛.
According to the systemic formalism, the basic function of MADH3 is described in the triad TSF as follows: (i) time and temporal facts are taken into account as qualitative and symbolic events described in a process ontology; (ii) space is associated with subcellular structures or compartments which are organised into a supramolecular-structure ontology; (iii) form and consequent changes (free, bound, chemically modified, etc.) are described through structure and/or process ontologies. This data organisation, which accounts for the activity of MADH3, is presented in Table 1. The first column shows the successive steps that delineate cellular states in signal transduction; the arrows indicate upstream and downstream events (open system). The second column refers to the ontology-type and the third one to vocabulary items; the GO numbers are indicated when available.
It must be stressed that such representation emphasizes the distinction, sometimes confused, between states (in our example, the first cellular state (i) consists of the combination of TSF dimensions) and stages (the first process stage consists of the transporters complex assembly, which says nothing about the structures involved 〚SARA:MADH3〛 or their location 〚cytoplasm〛).
4 Discussion and perspectives
To improve the design of information systems for the exploitation of genomic functional data, we must carefully discuss the concept of biological function and how this is represented in the form of computable symbols. This discussion must be developed from a thorough epistemological point of view. Until now, the variety and the multidimensional characteristics of the space–time and morphogenetic biological processes were poorly described, by linear and simplifying schemes, and these were used to propose softwares for data acquisition, analysis or modelling.
For this purpose, this epistemological point of view on the complex concept of biological function and on its modelling, led us to address (i) the nature of the biological functions as collections of organised/organising compounds, (ii) the way of describing them by instantiation within the systems that contain them, or by the causes and/or the finalities or the programs that cause their formation, (iii) the way they are acquired (emergence) by natural selection, dispositional regularities, relationships, etc., (iv) the actions that, in turn, trigger the system that formed them: positive and negative regulation, homeostasis, differentiation, etc.
This view favours systemic modelling 〚24, 25〛. Systemic modelling guides the modelling approach and the process of knowledge representation by asking the questions: to do what? where? why? how? These questions make it possible to understand the biological functions in the form of computable symbols by taking into account and modelling all features revealed by the former concepts on biological functions 〚6, 7〛. This enabled us to recognise the relevance of systemic modelling to account for the space–time and morphogenetic complexity of biological functions. The concept of general system developed in the systemic paradigm means that an effective instrumentation for the modelling of teleoadaptive (organised and organising) components in their environment can be developed, allowing us to establish a canonical model of the biological functions.
This is the first illustration related to the organisation of fields of knowledge. In biology, concept organisation, which is also known as ontological inquiry, is motivated by the need to design, to represent and to manage information, particularly structural and functional data. Ontology provides a model of the concepts in a given field and of the relationships among them 〚26〛. Recent studies pioneered by Karp 〚27〛 have produced a range of different results; they include the general Ontology for Molecular Biology (OMB) project 〚28〛, the Kyoto Encyclopedia of Genes and Genomes (KEGG) 〚29〛, the Gene Ontology (GO) project and ontologies dedicated to specific databases 〚30〛 and to annotation tasks 〚31〛.
From a biological perspective, the upper levels of abstraction ranged from (i) ‘Being’ 〚28〛 to (ii) ‘Genomic Object’ 〚31〛, including (iii) ‘Small Molecule and Macromolecular Metabolism’, ‘Structural elements’ and ‘Cell Process in EcoCyc’ 〚32〛, (iv) ‘Cellular Process, Cellular Function’ and ‘Cellular Component or Compartment’ in GO, (v) ‘Pathway, Gene and Molecule’ in KEGG and (vi) ‘Genetic Properties’, ‘Functional Properties’, ‘Post-translational Modifications’, ‘Cellular Role’, and ‘Subcellular Location’ in YPD 〚30〛; in addition, these top-level concepts were used to guide the creation of taxonomic hierarchies that describe classes of tangible (gene, molecule, etc.) and/or intangible (cellular role, subcellular location, processes, etc.) objects.
Some of the ontological studies mentioned above attempted to increase the consistency between databases. However, in the absence of epistemological thinking and of an explicit reference to a theoretical framework, it may be difficult to share data. For example, what links ‘Being’ (OMB) and ‘Structural elements’ (EcoCyc)? Is a ‘Cellular Function’ (GO) a ‘Cellular Role’ (YPD) or a ‘Structural Element’ (EcoCyc), or both? Are ‘Processes’ (GO) and ‘Pathways’ (KEGG) different? What does ‘function’ mean? What is the nature of links between ‘Function and Cell Processes’ (EcoCyc), ‘Compartment’ (GO), ‘Post-Translational Modifications’ (YPD)? How can the semantic confusion between a ‘Gene Product’ and its ‘Function’ be clarified? Which epistemic value distinguishes between ‘Local Function’ and ‘Integrated function’? 〚33〛. By providing us with the resources and the formalism of systemic modelling, the TSF triad provided a canonical representation of the biological function. This was done for TGFβ-dependent signalling and involved most of the concepts listed above. This model has an advantage thanks to the explicit reference to the well-established conceptual systemic paradigm and consequently, in the use of controlled vocabulary
Our accumulating experience with modelling will enable us to produce some kind of epistemological feedback. The representation of the biological functions was often exclusively perceived according to the modelling of circulating information as a flow; it has been postulated that (i) this information was a source of natural data, which could be represented as liquid flowing through pipes and (ii) this circulation does not affect the organisation (the network of pipes) in which it is exerted. Since Quastler’s pioneer texts 〚34〛, who interpreted the processes of biological interactions by use of the Shannon’s information theory, this metaphor guided the representation of the biological facts. This was inhibiting until the works of H. von Foerster 〚35〛 and Atlan 〚36〛. This interpretation can be widened appreciably (i) by taking into account both the information flow in the biological systems and its generation 〚37〛, (ii) by modelling the recursive formation process by which the system is organised and the changes that occur according to this information. This interpretation requires a more complex vision of the concept of function. To quote Valéry: “we reason only on models”; therefore it is essential to be attentive at the very beginning when designing and constructing the models before trying to apply computer programmes to unsophisticated and epistemologically contestable models. In the absence of this epistemological rigor, these interpretations and the relevance of such views would not be very valuable.
Based on this methodological approach, we can consider the interdependent concepts of information and organisation (and possibly of self-organisation) in ontological and phenomenological terms 〚38〛. Taking advantage of the modelling experience in progress as well as well as this internal epistemological criticism, we plan to concentrate our next efforts to examine the complex relationship between the generation, the circulation and the computation of the information on the one hand and between the formation of the organisation in these biological systems on the order hand; in addition, we should model this relationship (information–organisation–action–...〛. However, this complex organisation must be understood before we attempt to use a computer to solve a problem.
Acknowledgements
The authors would like to thank Charles Auffray, Bernard Pau and Franck Molina for stimulating discussions, and their colleagues at the first ‘Biosystemic Workshop’ held in Montpellier for their comments.
Version abrégée
La compréhension des processus biologiques passe aujourd’hui par l’interprétation des données du séquençage des génomes, ce qui rend nécessaire le développement de l’ingénierie des systèmes d’information dédiés à l’exploitation des données fonctionnelles. Une des questions de base qui se posent lors des premières étapes de la conception de tels systèmes d’information concerne le type des informations à traiter, leurs propriétés et leurs relations, ce qui nécessite un examen attentif des concepts de gène et de fonction. Actuellement, les représentations de la notion de fonction de gène s’appuient largement sur le modèle de Beaddle et Tatum : un gène, un enzyme ou sa variante : un gène, un peptide. Toutefois, ces modèles « particulaires », qu’on serait assez tenté de qualifier de statiques, ont de plus en plus de difficultés à rendre compte de la plasticité des activités exercées par les gènes et/ou leurs produits. Un modèle « constructiviste » du gène a été proposé récemment, qui reprend l’ancienne conception de R. Godlschmidt d’un gène indissociable de son action et du contexte dans lequel celle-ci s’exerce. Cette interprétation récursive du gène et de sa fonction entraîne des conséquences majeures sur leur représentation : la fonction n’est pas un attribut supplémentaire du gène ; au contraire, elle « décrit » le gène perçu comme un objet actif.
Le paradigme systémique propose un cadre théorique argumenté pour la représentation d’un objet actif. Il permet en effet de rendre compte du caractère indissociable de l’activité d’un composant (ou d’un ensemble de composants, c’est-à-dire d’un système) et de son évolution dans un environnement par rapport aux finalités auxquelles il est associé (la ou les fonctions qu’il exerce). La notion de paradigme est entendue au sens de E. Morin et « contient l’ensemble des concepts et des catégories majeurs ainsi que le type de relations d’attractions/répulsions (conjonction, disjonction, implication…) entre ces concepts et catégories… un paradigme n’explique pas mais il permet (…la modélisation) de l’explication ».
Le concept de base du paradigme systémique est l’action et l’axiomatique de la logique conjonctive, basée sur les principes de synchronicité, diachronicité et récursivité, permet d’assurer l’instrumentation modélisatrice de la systémique.
- • le principe de synchronicité rend compte des comportements du système au sein de son environnement (le fonctionnement dans le contexte) ;
- • le principe de diachronicité rend compte des transformations endogènes du système au fil du temps ;
- • le principe de récursivité rend compte des interactions du système et de ses finalités, de l’action et de ses résultats (les résultats de l’action sont nécessaires à l’action qui les génère ; autrement dit, un système engendre ses finalités en fonctionnant).
L’idéogramme de système général rend compte des cinq concepts majeurs du paradigme systémique : un composant ou un ensemble de composants (système) évoluant téléologiquement (finalité) dans un environnement (contexte) et qui se transforme (évolution) en fonctionnant.
Action et fonction se définissent récursivement (les résultats de l’action – ce qui est fait, la fonction – étant indissociables de l’action qui les produit) et sont incluses dans le concept de processus, lui-même décrit par son exercice et son résultat : un processus peut être représenté par le déplacement d’un objet identifiable dans un référentiel temps–espace–forme (TEF) ; autrement dit, un processus résulte de la conjonction d’un transfert spatio-temporel et d’une différenciation morphologique. Le concept de forme est utilisé ici pour décrire ces entités à la fois organisantes et organisées, par lesquelles se manifestent les processus. Il apparaît ainsi qu’un processus au niveau n pourra être représenté par son résultat au niveau n + 1, ce qui s’exprime par l’introduction du concept de processeur. Un processeur sera symbolisé par une boîte noire dont on pourra à chaque instant décrire l’état. L’articulation des trois catégories prototypiques de processus 〚(i) morpho-différentiel : transformation, différenciation..., (ii) transfert spatial : transport, transmission..., (iii) transfert temporel : stockage, mémorisation...〛 au sein d’une architecture de processeurs assure la représentation d’organisations hiérarchiques et/ou emboîtées.
L’intelligibilité des fonctions biologiques s’exprime par des modifications irréversibles dans le temps, portant sur la forme (par exemple, l’activation d’une molécule par phosphorylation) et la localisation (par exemple, le compartiment subcellulaire dans lequel intervient telle fonction) d’entités biologiques. Ces changements sont associés récursivement aux modifications de l’environnement. Ainsi, un processus de signalisation intracellulaire (par exemple, la voie de signalisation dépendante du TGFβ) sera décrit par l’articulation des trois catégories de processus mentionnées précédemment, au sein d’une architecture de processeurs biologiques (bioprocesseurs) qui correspondent aux complexes moléculaires. Ces complexes moléculaires résultent, pour leur part, de l’exécution de processus d’interaction moléculaire à des niveaux hiérarchiques inférieurs. Inversement, au niveau organismique (par exemple, au cours des étapes précoces du développement embryonnaire), la voie de signalisation dépendante du TGFβ constitue, ainsi que d’autres voies de signalisation (notamment, la voie VCC/β-caténine), un élément du processus de « ventralisation » de l’embryon et, comme tel, pourra être modélisé par un processeur.
La référence explicite au paradigme systémique introduit une cohérence forte dans la représentation des fonctions biologiques, puisqu’un même schéma va s’appliquer à la modélisation des processus, que ceux-ci interviennent aux niveaux moléculaire, cellulaire ou organismique. La notion de finalisation, qui est centrale dans le paradigme systémique, est prise dans son sens téléonomique, porteur d’une forte valeur heuristique. En outre, le formalisme systémique permet de restituer les aspects dynamiques du fonctionnement des systèmes. En effet, la représentation courante des processus biologiques correspond à une vision cinématique qui ordonne une succession temporelle d’états. Ceci est différent d’une vision dynamique, qui intègre les événements qui président à ces changements. En décrivant un état par la conjonction de changements spatio-temporels et morpho-différentiels, le formalisme systémique permet une représentation dynamique d’un processus, en renseignant sur les états d’un système et son évolution. Un exemple est présenté pour illustrer la pertinence de la modélisation systémique pour la représentation et l’exploitation des données de la génomique fonctionnelle. Cet exemple porte sur la conception d’un vocabulaire contrôlé dédié à la description et à la qualification des fonctions biologiques (organisation des données rendant compte de l’activité du gène SMAD3/MADH3).
Le formalisme systémique conduit à interpréter le traitement des processus moléculaires en termes d’opérations organisationnelles produisant et transformant l’information génétique et à approfondir l’intelligibilité de la génération, de la circulation et du calcul de l’information dans la mise en place des organisations biologiques.



