Every conservation project needs to answer different questions about the organisms under investigation. Their systematics, phylogenetic relationships and the relatedness among individuals are only some of the different aspects of fundamental importance in planning the conservation of the biodiversity of a particular area. Moreover, it is always more urgent to start captive breeding projects to manage the last individuals of an endangered species. Therefore, conservation genetics has became one of the most important aspect in the conservation of the biodiversity and its management.
Several kind of molecular markers are available but none of them are useful for all applications [1]. Most of the systematics and phylogenetics studies are approached with mitochondrial DNA sequencing. However, mitochondrial DNA phylogeny represents only the genealogy of a single gene that is almost only maternally inherited. Therefore, the results need to be confirmed by congruence with the results obtained from a nuclear gene.
Population genetics, hybridization and paternity analysis use another group of molecular markers, the microsatellites. These molecular markers are short tandem repeated sequences that, due to their high mutation rate, are variable enough to discriminate at the individual level. However, the isolation of species-specific microsatellite loci still requires a certain amount of laboratory work and resources that are not always available for all laboratories. Cross-specific amplifications of microsatellites could be a short-cut method to overcome this limitation but this strategy does not always work properly and often does not work at all.
A new molecular technique called Amplified Fragment Length Polymorphism (AFLP) was introduced by Vos and collegues in 1995 [2]. AFLP seems to be promising in providing a very large number of polymorphic markers with a very fast and relative simple laboratory work [3,4]. This technique seems to be very robust and its reproducibility was tested among different laboratories with high success. Another important aspect of this technique is that it does not require any prior knowledge about the taxa that is going to be investigated, and the same protocol could be used for a wide range of organisms.
In this study we investigate the usefulness of the AFLP technique applied to three different conservation aspects. The first is a phylogeographical study of the wild populations of Bearded pig from three localities of Southeast Asia: Malaysia, Borneo and Sumatra. The second is a study to detect hybridization among Gallus species reared in captivity and the third is a study of paternity analysis and sex determination in parrots. We compared the results obtained using AFLP with those obtained from mitochondrial DNA sequencing, microsatellites analysis and other well known molecular techniques.
All the AFLP analysis that we used followed the protocols described in the paper by Vos and colleagues [2]. We used the restriction enzymes EcorI and TaqI and a combination of five primers forward and seven primers reverse. The AFLP fragments were run on an automatic sequencer 3100 ABI and analyzed using the software Genotyper v3.7 (ABI).
1 Phylogeography of the Bearded pig
The bearded pig (Sus barbatus) is distributed across the southeast Asia, occurring in the Malaysia peninsular, Sumatra, Borneo, Palawan and neighboring island in the west Philippines. Actually, three subspecies are recognized: S.b. barbatus on Borneo island, S.b. oi in the peninsular Malaysia and Sumatra and S.b. ahoenobarbus on Palawan and Philippines islands. We have investigated its phylogeography using a mitochondrial DNA phylogeny obtained from the sequencing of the entire cytocrome b and the first part of the control region for a total of 1740 nucleotides (Fig. 1a). We have analyzed eleven samples from Borneo, eighteen from Malaysia and five from Sumatra. The neighbour-joining analysis [5] of the pairwise distance matrix obtained using the Tamura and Nei method [6] showed that the samples from Sumatra are very divergent respect to those of Borneo and Malaysia peninsula that, instead, result grouped in the same cluster with no clear geographical differentiation. Using five AFLP primer combinations we obtained 83 polymorphic bands that were analyzed by a principal component analysis. The AFLP results (Fig. 1b) confirmed the high divergence of the sumatran samples from Borneo and peninsula Malaysia and, also, separated the group of Bornean bearded pigs from those of the peninsular Malaysia.
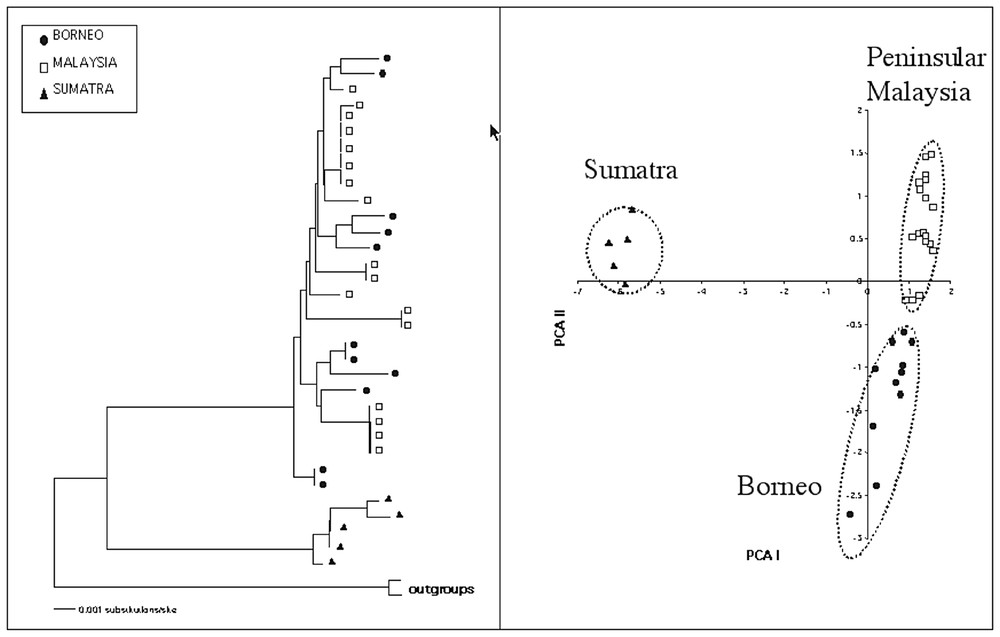
In this figure are represented the results of the analysis of 33 berded pigs from Borneo (fill circle), Peninsular Malaysia (open square) and Sumatra (filled triangle). (Left panel) Neighbor-joining tree computed using a Tamura & Nei (1993) distance matrix of 1740 nucleotides of mitochondrial DNA, including the entire cytochrome b and the left part of the control region. (Right panel) A principal component analysis of 83 polymorphic bands obtained using AFLP technique.
2 Detect hybridization in Gallus
The genus Gallus comprises four species: the red junglefowl (Gallus gallus), distributed from NE India throught Bangladesh, Nepal, Thailand, peninsular Malaysia and N Sumatra; the grey junglefowl (Gallus sonnerati), distributed in peninsular India; the ceylon junglefowl (Gallus lafayetii), distributed only in Sri Lanka; the green junglefowl (Gallus varius), distributed in Java and the neighbouring islands. One of the main threat for the conservation of this genus is the hybridization with domestic fowls. Therefore the aim of this study was to find a method to identify hybrids. The study is based on a sample of forty-six fowls including all the four species plus three known hybrids between the red junglefowl and the grey and green junglefowls. We initially used a phylogenetic approach using the sequencing of the first hypervariable part of the mitochondrial control region. After that, we analysed the samples using fifteen microsatellites and conducted a principal component analysis of the individual genotypes (Fig. 2a). To evaluate the usefulness of the AFLP markers compared to sequencing and microsatellites analysis in detecting the known hybrids we used eight primer combinations for a total of 84 polymorphic bands. From the principal component analysis of the AFLP data the four species resulted well separated and the three hybrids were correctly identified (Fig. 2b).
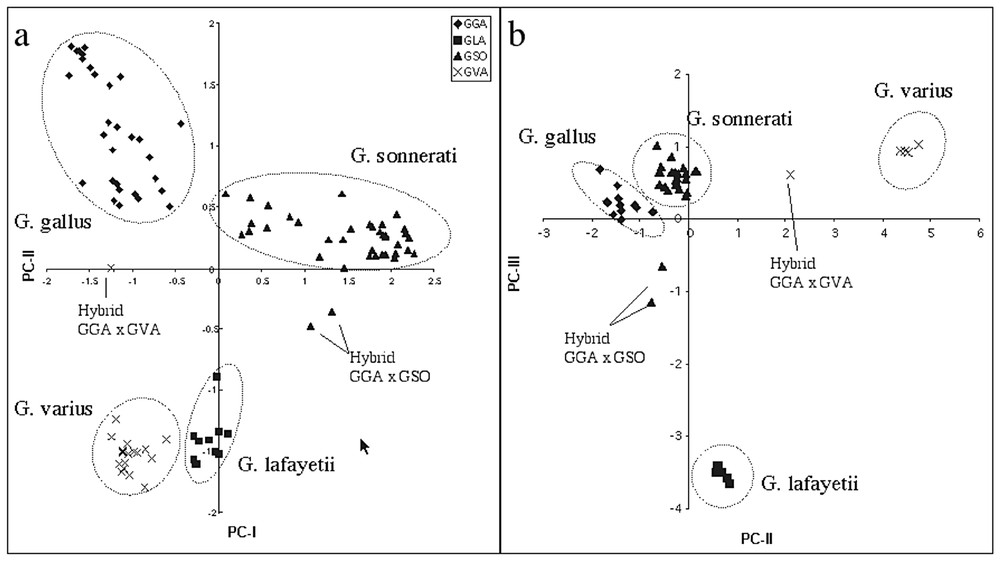
Principal component analysis of the four species of fowls and three hybrids, obtained using fifteen microsatellite (a) and eighty-four AFLP polymorphic bands (b).
3 Paternity analysis and sex determination in parrots reared in captivity
Parrots are one of the most threatened taxon among birds because of the disappearance of their natural forests and, also, because they are subject to illegal trades and captive breeding activities. The CITES authorities often require the genetic analysis to confirm the declarated captive origin of the birds possessed by breeders. To do parental analysis it is necessary to study a high number of molecular markers variable enough to discriminate among individuals whose variability is inherited in a mendelian fashion. We have tested the AFLP technique and compared the results with a minisatellite fingerprinting analysis that was used in our laboratory. The AFLP showed several polymorphic bands that were all inherited according to the Mendelian model (Fig. 3). Variability was very low in several species, probably due to the high level of imbreeding present in the breeding stocks. However, using the AFLP we could confirm a case of wrong parental analysis previously found using the minisatellite fingerprinting technique (Fig. 4).

Parental analysis of a family of Ara ararauna obtained using the AFLP. The polymorphic bands are indicated by a letter and their size.
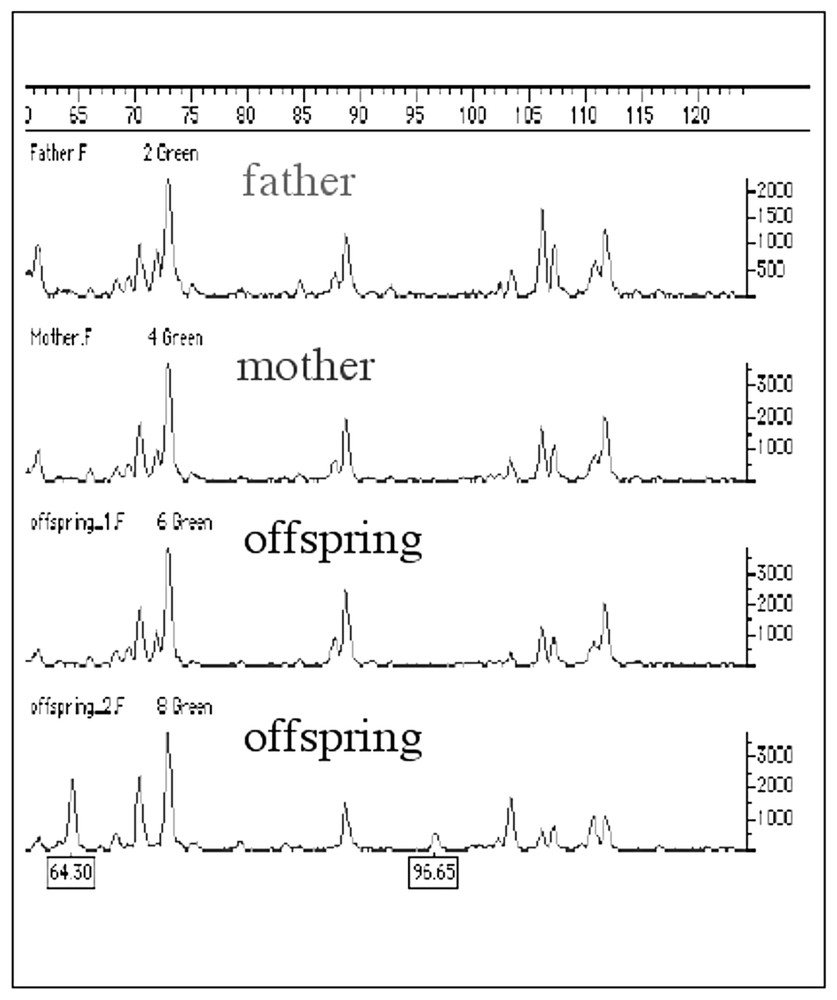
Diagnosis of parentage exclusion in a putative family group of Parrots. The offspring at the bottom cannot be an offspring of the two putative parents since it has two bands not in common with them.
By using the AFLP techniques it was also possible to identify two bands associated to the sexual chromosomes that are useful for sex determinations in the genera Ara and Anodorhynchus (Fig. 5).
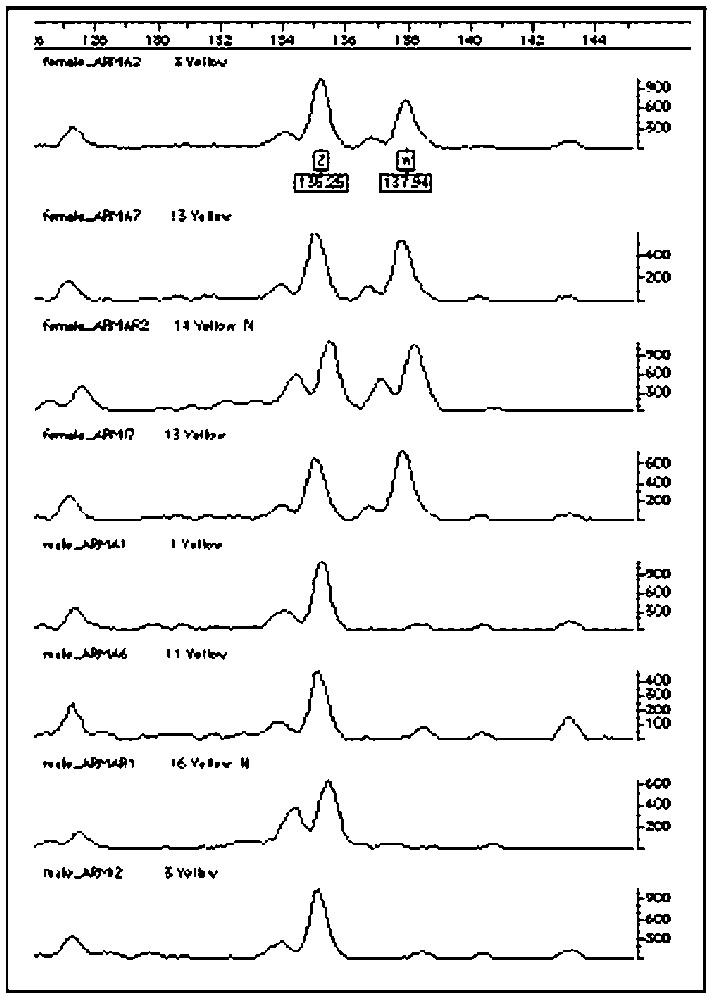
In this figure are showed two bands obtained by AFLP analysis that identify the sex in three species of the Genus Ara. The females have two bands that represent both sexual chromosomes (Z and W) while the male present only a single band (ZZ).
4 Conclusion
In conclusion, the AFLP technique has been shown to be a very good molecular technique for biodiversity conservation and management studies and will be useful in a wide range of applications.



