1 Introduction
The European mink Mustela lutreola (L., 1761) is one of Europe's most endangered carnivores, since recent population censuses estimate that it now exists in isolated populations covering only one-fifth of its former range [1]. The main population subsists in the territory of the former Soviet Union and isolated populations exist in Spain and France. The western population is particularly threatened and is declining in over 50% of its range [2]. The drastic decline of effectives affords the problem of reduction of the genetic diversity. We chose to characterise microsatellite loci in order to investigate the pattern of decline of this species. Nevertheless no primer pairs allowing microsatellite characterisation have been developed for European mink. Such a development requires the construction and screening of a partial genomic library and the sequencing of clones. The cost of this step was out of range for us so we decided to exploit sequence conservation and to work with primers developed for related species.
We used a widespread species Mustela putorius (European polecat) relative to Mustela lutreola as a “standard” to identify alleles and to evaluate the potential polymorphism at each locus. For such a purpose 69 road-killed individuals of Mustela putorius, were collected in 4 regions in France (Fig. 1).
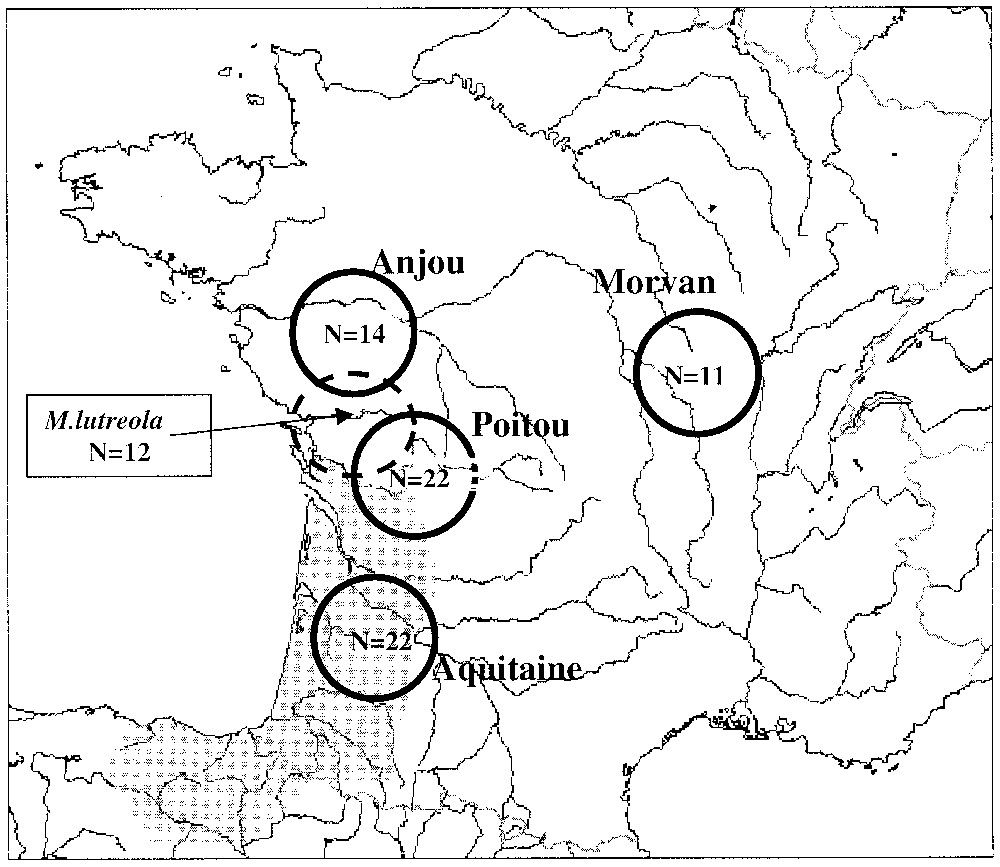
Polecat sample areas (in gray, current distribution of European mink western population, N=number of sample).
Finally, we genotyped 12 individuals of European mink using 8 primer pairs and we compared the result with those characterising the polecat.
2 Materials and methods
Previous studies [3,4] described 16 primer pairs useful for Mustela vison genotyping. Moreover the screening of database (Genbank) allowed us to find two other M. vison microsatellite loci (GB: U92534 Brusgaard, 1997; GB: AY040547, Vincent et al., 2001) and we evidenced a simple sequence repeat (SSR) in the coding sequence of Mustela putorius furo Kv1.4 ventricular voltage potassium channel (FK1) (GB: U06156.1). In total, 19 primer pairs were used for this work.
3 Results and discussion
Nineteen primer pairs originally developed for genotyping American mink were assayed. Eight of these allowed us to characterise polecat and European mink individuals (Table 1). We developed one primer pair to amplify the SSR evidenced in Mustela putorius furo Kv1.4 FK1 coding region. Two primer pairs assayed did not allow us to obtain amplification. One can make the hypothesis that mutation occurred in the primer fixation region leading to unamplification of allele. The low ratio of such result (2/18) illustrates the relatedness of American mink with polecat and European mink. We were unable to identify alleles with 8 primer pairs. The separation of amplification product following PCR with a microsatellite locus derived primer pair reveals complex pattern as several bands were present. The identification of microsatellite allele could be difficult as the various mechanisms of SSR evolution (strand slippage, unequal crossing over) lead to variation in the number of repeats. Moreover the high level of homozygosity expected for endangered species and the low number of samples could compromise locus identification. If there is no polymorphism at the SSR locus, it is impossible without sequencing to identify the band corresponding to the microsatellite allele. One could imagine to score such a locus as monomorphic but mutation in the primer site or deletion could have occurred leading to nul allele which will be mis-scored as monomorphic, causing an underestimation of genetic diversity. The Table 2 summarises the range of allele size determined for M. vison, M. putorius and M. lutreola. Except for locus Mvi1843, allele size ranged closely to the value determined for M. vison. Thus the conservation of loci seems to be effective for the primer pair used.
Results obtained for European mink from 18 primer pairs designed for American mink microsatellite loci genotyping
| Primer pair origin | O'Connel et al. | Fleming et al. | Vincent et al. | Brusgaard |
| Number of primer pair | 9 | 7 | 1 | 1 |
| No amplification | 2 | 0 | 0 | 0 |
| Unable to identify alleles | 3 | 5 | 0 | 0 |
| Genotyping | 4 | 2 | 1 | 1 |
Comparison of allele size revealed for M. vison, M. lutreola, M. putorius. For M. vison original allele size deduced from sequencing, for M. putorius and M. lutreola range of allele size evidenced in this study
| Locus | Mvi002 | Mvi020 | Mvi072 | Mvi389 | Mvi1843 | Mvi054 | Mvi111 | PutFK1 |
| M. vison | 181 | 155 | 243 | 103 | 147 | 118 | 89 | 151 |
| M. putorius | 188–204 | 137–139 | 248–256 | 103–107 | 118 | 106–118 | 89–107 | 145–151 |
| M. lutreola | 192 | 137 | 252–260 | 103 | 130–132 | 118 | 89 | 149–151 |
Nevertheless the exploitation of sequence conservation could lead to bias concerning the estimation of genetic diversity. Several indications have proved that SSR can cause quantitative changes in gene expression and function and by the way affect phenotype [5]. If a sequence is conserved from one species to another, we can imagine that this conservation could be partially due to the fitness of the microsatellite locus. In order to investigate this point, we analysed the distribution of allele size for European mink and polecat. Under a random model of evolution we expect roughly a normal distribution of alleles.
Locus Mvi002 presented a high frequency for a 188 pb allele and exhibited a roughly normal distribution for alleles ranging from 196 pb to 204 pb (Fig. 2). One should suspect a positive effect on fitness of 188 bp allele; nevertheless a search on Genbank did not reveal significant homology with a gene.

Distribution of allele frequency at locus Mvi002.
At locus Mvi111 (Fig. 3) the distribution of allele size in polecat is normal and European mink appears fixed for a smaller allele. Four polecat individuals showed the allele fixed in European mink. The clear cut variation in allele size between the two species (5 repeats; 10 base pairs) allows us to exclude that the European mink allele could originate in the polecat by replication slippage. We can therefore hypothesise that a hybridisation event occurred. Forming a syngameon according to the Templeton's definition [6], European polecats, Steppe polecats and European minks are able to hybridise and produce fertile hybrids [7]. But in nature such hybridisations were only suspected through morphological identification. We present here the first element allowing to support the evidence that interspecific hybridisation occurred between polecat and European mink in natural habitats.

Distribution of allele frequency at locus Mvi111.
At locus Mvi054, European mink were fixed for a 118 pb allele. Except 7 individuals out of 69, polecats are fixed for a 114 pb allele. Three polecats exhibited 118 pb allele. As for locus Mvi111 we can hypothesise a hybridisation event between polecat and European mink. Nevertheless the small difference in allele size could be related to mutation by random slippage. Moreover the fact that this locus appears fixed in polecat contrary to locus Mvi111 addresses the problem of the possibility of variation at this particular locus. Searching on Genbank showed that locus Mvi111 did not reveal significant homology with other sequence but Mvi054 presents homology with Felis catus sequence (Fasta score 1.3×10−6). Although no function is affected today to this sequence, it could be related to a gene and therefore subjected to selection pressure.
The same situation was found for locus Mvi389 where polecats were fixed for a 107 pb allele, except two individuals exhibiting the 103 pb allele fixed in European mink samples. Finally 4 loci (Mvi389, Mvi020, Mvi054, Mvi1843) exhibited a low level of allelic variation with two main alleles roughly fixed one in each species. The four other loci (Mvi002, Mvi072, Mvi111, PutK1) showed a higher allelic diversity and moreover the frequency of alleles was approximately normally distributed. These examples emphasise the need of a careful analysis for microsatellite data. According to the locus the level of information could greatly vary.
Finally sequence conservation allowed us to genotype 69 polecats and 12 European mink. The Fis value, as measured by the Genetix program, are summarised in Table 3. The Morvan population exhibited a low Fis value. By contrast the Western France population and European mink showed a high Fis value. This emphasises the conservation problem for these species in this area. One can address the question whether the same causes affect both species decline.
Variations of the index Fis among four polecat populations and European mink western population from France
| Population | Anjou | Poitiers | Aquitaine | Morvan | M. lutreola |
| N=14 | N=22 | N=22 | N=11 | N=12 | |
| F is | 0.1389 | 0.2416 | 0.1502 | 0.0429 | 0.1907 |
Although a bottleneck may chiefly affect the number of polymorphic loci [8], the heterozygosity level in European mink remained clearly lower than that observed in polecats. Mink population showed a significant heterozygote deficit revealing that inbreeding affects populations. Species near extinction generally exhibit both a decline in their range and depletion in genetic diversity (cf. [9]). The incidence of loss of genetic diversity increasing the vulnerability of population in changing environment is often be underestimated but such a loss of genetic variability could be regarded as a major threat for European mink conservation.



