1 Introduction
Classical Science, that is the scientific activities that have sprung up since the days of Descartes, Newton and Leibniz, has always been in search for simplicity. More explicitly, it is based upon four ‘principles’ expressed by Descartes in the Discours de la Méthode [1]. These ‘principles’ are expressed below.
Le premier était de ne recevoir jamais aucune chose pour vraie, que je ne la connusse évidemment être telle : c'est-à-dire d'éviter soigneusement la précipitation et la prévention ; et de ne comprendre rien de plus en mes jugements, que ce qui se présenterait si clairement et si distinctement à mon esprit, que je n'eusse aucune occasion de le mettre en doute.
Le second, de diviser chacune des difficultés que j'examinerais, en autant de parcelles qu'il se pourrait, et qu'il serait requis pour les mieux résoudre.
Le troisième, de conduire par ordre mes pensées, en commençant par les objets les plus simples et les plus aisés à connaı̂tre, pour monter peu à peu, comme par degrés, jusques à la connaissance des plus composés ; et supposant même de l'ordre entre ceux qui ne se précèdent point naturellement les uns les autres.
Et le dernier, de faire partout des dénombrements si entiers, et des revues si générales, que je fusse assuré de ne rien omettre.
The second ‘principle’ is no doubt reductionist in its essence and, for this reason, adverse to present studies on complex systems. In spite of the fact that these analytic and reductionist approaches have led to important discoveries, there is little doubt that the World is complex in its essence and cannot be correctly understood through the classical reductionist approach. This is precisely what has recently become evident in different fields of Science ranging from fundamental Physics to Social Sciences [2–8]. It is clear that the living systems on Earth are complex systems and that it is hopeless to try to understand “what is life” through the sole study of the macromolecules that make up the substance of these systems.
The aim of the present contribution is threefold:
- – first, to present a tentative definition of reduction, integration, emergence and complexity;
- – second, to give a brief overview of the main features of complex systems;
- – third, to discuss more thoroughly the most important of these features, the one which is at the very basis of the concept of complexity, namely information.
Although this discussion will be general, it is more specifically oriented towards dynamic biological systems.
2 A tentative definition of reduction, integration, emergence and complexity
From a philosophical viewpoint, the term of reduction can, at least, have two different meanings. It may refer to the mental process of deriving a scientific theory from another one, more general and embracing [9,10]. In this perspective, a biological theory, for instance, could be reduced to a more general physical one. If such a reduction could be pursued ad infinitum, this would imply unity of Science.
But there is a second type of reduction that is directly related to the problem of emergence and complexity. Let us consider a system made up, as any system, of a number of sub-systems. One can expect three types of situations to occur. The overall system and the set of the isolated component sub-systems have the same degrees of freedom, or the same entropy, or the same properties. Then, the overall system is not a real one, but the simple union of the two component sub-sets. The second possible situation occurs when the overall system has less degrees of freedom, or smaller entropy, than the set of the component sub-systems. It therefore behaves as a real system, for it displays some sort of integration of its elements as a coherent whole. The integrated system therefore displays collective properties distinct from those of the isolated component sub-systems. The last situation is observed if the overall system has more degrees of freedom, or more entropy, than the set of the component sub-systems. One may expect this system to have more wealthy collective properties than the integrated system considered above. It can be defined as an emergent, or complex, system. It results from this definition that neither the properties of an integrated system, nor those of a complex system can be reduced to the properties of their component sub-systems.
As the concepts of integration and emergence are related to that of information, it is thus clear that the analysis and discussion of this concept should be central in any discussion on complexity.
3 Main features of complex systems
Before discussing more thoroughly the logical foundations of the concepts of information, integration and emergence, it is of interest to present briefly the main features of complex systems [7]:
- – a complex system should possess information; this important matter will be discussed later on;
- – a complex system exhibits a certain degree of order; it is neither strictly ordered, nor fully disordered;
- – complex systems should display collective properties, that are properties different from those of the component sub-systems;
- – a complex system is usually not in thermodynamic equilibrium; it is thermodynamically open and displays non-linear effects;
- – a complex system is in a dynamic state and has a history; this means that the present behaviour of the system is in part determined by its past behaviour;
- – a complex system has emergent collective properties.
As far as we know, it seems that properties 1, 2 and 6 are mandatory for the definition of a complex system.
4 Information, integration and emergence in biological complex networks
Total information of the living cell is often identified to genetic information. This is too restrictive a view of the concept of information. As a matter of fact, one may expect most biological networks to store information for they associate in space and time, according to a strict order, many chemical reagents. Classical Shannon's communication theory [11–14], however, is not ideally suited for describing what network information is. It has therefore to be revisited and altered in order to allow this study. Moreover, many papers have recently been devoted to networks, in particular to metabolic networks [15–19]. Before studying integration and emergence, one has to present a brief overview of what a biological network is.
4.1 Biological networks
A network is a set of nodes connected according to a certain topology. In a metabolic network, for instance, the nodes are the various metabolites, or the enzyme–metabolite(s) complexes, and the edges that connect the nodes the corresponding chemical reaction steps. Networks belong to different types known as random, regular, small-world and scale-free [15]. In random graphs, the nodes are mutually connected in a random manner according to a Poisson distribution, whereas in regular networks, node connection is effected through a fixed topological rule. Small-world networks display a fuzzy topology, midway between pure randomness and strict regularity. Sale-free graphs display both poorly and highly connected nodes.
A network is characterized by a parameter called diameter. If we consider all the possible pairs of nodes in a graph and the shortest distance, that is the smallest number of steps within a pair, the network diameter is the mean of the shortest distances. The immediate consequence of this definition is the increase of the diameter of a random graph as the number of nodes increases.
Metabolic networks possess an interesting property that is worth discussing briefly here [18]. Sequencing genomes of very different organisms, ranging from mycoplasms and bacteria to man, have allowed us to know most of the enzymes present in these organisms, and therefore most of the reactions they catalyse. As a consequence, this allows us to construct, in principle, the metabolic networks of these living systems. Depending on they are primitive or evolved, the number of nodes of their metabolic graphs are very different. Still, the graph diameter remains nearly constant along the phylogenetic tree. The reason for this constancy is that metabolic networks are not random but possess a fuzzy organization. They are of the small-world type, and more and more connected as the organism is more evolved [18,19].
4.2 Subadditivity principle, reduction and integration
Let us consider a composite dynamic network made up of four classes of nodes. A first class, X, collects nodes associated with a discrete variable xi. A second class, Y, describes a different property associated with a different discontinuous variable yj. A third one, XY, has both properties xi and yj, and a last one has none of them. Each node of X and Y has a certain probability of occurrence, p(xi) and p(yj), respectively. Similarly, each node of the class XY has a probability p(xi,yj). If the events xi and yi are independent, then:
| (1) |
| (2) |
The concept of information is related to that of uncertainty. The larger the uncertainty of a message and the larger is its information content. Put in other words, the information of a system is its ability to perform a difficult, and therefore improbable, task. The higher this difficulty, and the larger is the information required to perform the task. Therefore, the essence of information should be related to that of uncertainty. Any mathematical function aimed at measuring the degree of uncertainty should meet two axioms: that of monotonicity and that of additivity. Monotonicity means that the uncertainty should increase regularly with the number of states xi or yj. Additivity expresses the view that if the discrete variables X and Y are independent, the uncertainty of the couples XY should be equal to the sum of the uncertainties of X and Y. The only simple function of a probability pi that meets these two requirements is:
| (3) |
| (4) |
| (6) |
| (7) |
Now, if the discrete variables X and Y are correlated and if the sum of the probabilities, p(xi),p(yj), and p(xi,yj), are all equal to unity, then one can demonstrate that the mean value of the conditional probabilities is larger than the mean of the corresponding probabilities. This implies that:
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
Information is therefore the piece of entropy that should be added to the joint entropy H(X,Y) in order to obtain the sum of individual entropies H(X) and H(Y). If the subadditivity principle applies, information is positive or nil, but if it does not, that is if
| (13) |
4.3 Integration and emergence in uncorrelated networks
The above reasoning, and the subadditivity principle which is at the heart of conventional Shannon's communication theory, rely upon the view that variables are correlated. If, however, these variables are not statistically correlated, but represent events that physically interact, the concept of information can still be applied to this situation. To illustrate this idea, let us consider a simple enzyme-catalysed chemical reaction AX+B→A+XB. If the reaction is catalysed by enzyme E, this may imply that AX and B can form binary and ternary complexes with the enzyme E, as shown in Fig. 1. Catalysis takes place on the ternary E·AX·B complex, which decomposes, thus leading to the regeneration of the free enzyme and to the appearance of the reaction products A and XB.
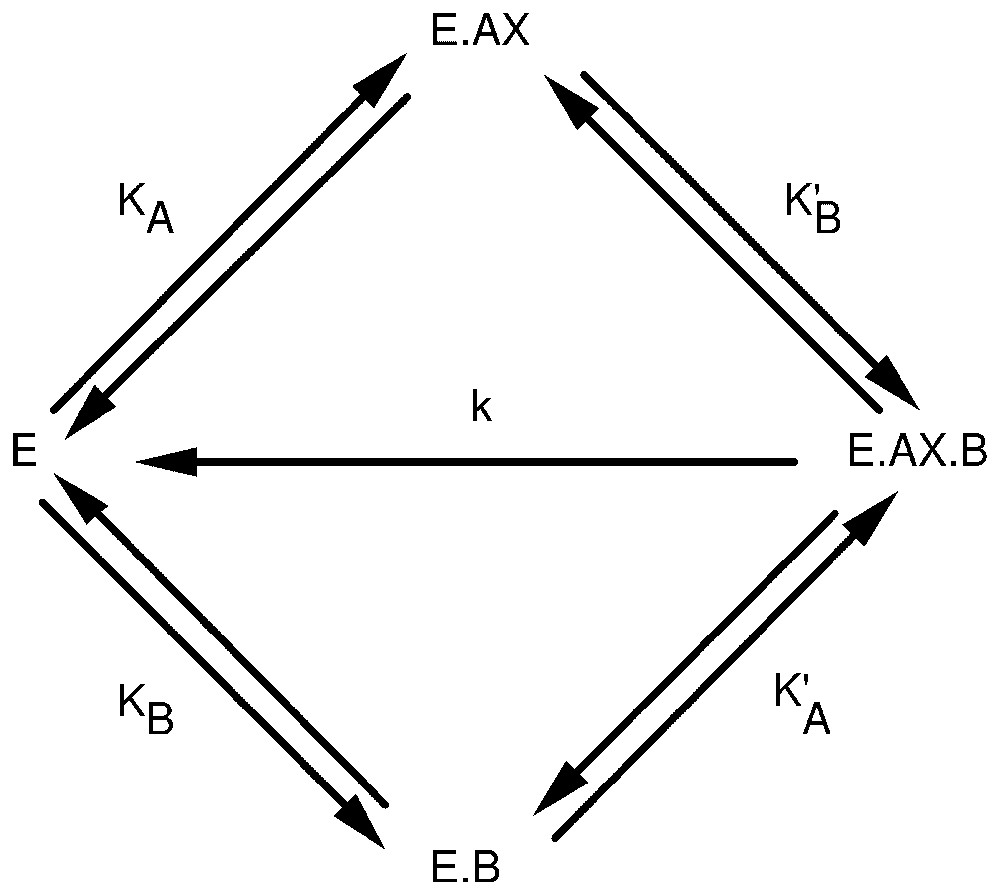
Random addition of substrates, AX and B, to an enzyme E. KA, K′A, KB, and K′B are the apparent binding constants of the substrates to the enzyme. k is the catalytic constant.
Let us assume for the moment that the overall system is close to thermodynamic equilibrium, which implies that the rate constant of decomposition of the ternary complex E·AX·B is small relative to the other rate constants. Then, one can easily derive the expression of the probabilities for the enzyme to bind AX, p(AX), or to bind B, p(B), or to bind both AX and B, p(AX,B). Similarly, one can derive the expression of conditional probabilities, p(AX/B) and p(B/AX). If the concentrations of AX and B are constant in the reaction mixture, the probabilities p(AX) and p(B) are not correlated. As a matter of fact, each of them assumes one fixed value only. But one can still extend the concepts of entropy and information to the present situation by setting: (14)
These expressions are indeed identical to equations (5) and (6), if X, Y and XY each assume one value only. Expressions of the probabilities that appear in equations (14) are dependent on the concentrations of the reagents, AX and B, as well as upon the affinity constants KA, K′A, KB and K′B. One can demonstrate that if KA=K′A, which implies from thermodynamics that KB=K′B, then (15)
| (16) |
Alternatively, if KA<K′A and KB<K′B, then (17)
| (18) |
The system behaves as an integrated coherent entity. Its entropy, H(AX,B), is smaller than the sum of the entropies, H(AX) and H(B), of the two component sub-systems. The information of the network, I(AX:B), is thus positive and equal to the extent of the integration process.
The last case is even more interesting. If KA>K′A and KB>K′B, then (19)
| (20) |
As the subadditivity principle is at the root of standard communication theory, the last conclusion above is at variance with this theory. It is therefore of interest to make clear the reasons for this disagreement. Classical Shannon's communication theory is based on two different implicit postulates: the existence of a statistical correlation between discrete variables and the assumption that the sums of the probabilities of occurrence of the corresponding states are all equal to unity. In the case of a simple enzyme network, as the one shown in Fig. 1, none of these conditions is fulfilled. Therefore, for an enzyme reaction, subadditivity may, or may not, apply and one can also reach the same conclusion for metabolic networks. As a matter of fact, for the enzyme process of Fig. 1, if K′A>KA and K′B>KB, the binding of B to the enzyme facilitates that of AX, and conversely. Subadditivity should apply to this situation. But if KA>K′A and KB>K′B, then the binding of B hinders that of AX, and conversely. In this case, subadditivity does not apply anymore. Therefore, in the present case, positive or negative information does not originate from statistical correlation between discrete variables, but from the physical interaction between two binding processes.
It is of interest to stress that, if subadditivity does not apply, this means that the probability of occurrence of the E·AX·B complex is smaller than the product of the corresponding probabilities of occurrence of the binary complexes E·AX and E·B. This implies in turn that the ternary complex E·AX·B is destabilised relative to the binary complexes E·AX and E·B. One can see that, under these conditions, the Michaelis constants for the two substrates AX and B as well as the catalytic constant are increased. Therefore, the intrinsic properties of the enzyme network are altered owing to the high information content of the E·AX·B complex. The situation that is expected to occur is just the opposite if there is less information in the E·AX·B complex than in the set of the E·AX and E·B binary complexes.
Thus far, the simple enzyme network was assumed to occur under quasi-equilibrium conditions. If, however, the system departs from this state, that is if the catalytic rate constant, k, assumes rather high values, the system can possibly reach a steady state. Under these conditions, the probabilities for the enzyme to bind AX, B, or both AX and B depend also upon the k value. As a matter of fact, the probabilities p(AX), p(B) and p(AX,B) decrease as k is increased. However, the decrease of p(AX,B) is steeper than that of p(AX) and of p(B), in such a way that H(AX,B) becomes larger than the sum of H(AX) and of H(B). This means that, as the system departs from its initial pseudo-equilibrium conditions and becomes more ‘open’, it tends to generate negative information. Moreover, this situation takes place even if KA=K′A and KB=K′B, that is even if the binding of the two substrates to the enzyme do not physically interact.
5 General conclusions
Classical Shannon's theory is aimed at deciphering how information is conveyed within a channel from a source to a destination. The basic principles and the mathematical formulation of this communication theory, however, have to be revisited and altered as to offer a clear-cut and mathematically sound definition of the concepts of reduction, integration, emergence and complexity in biological systems. The reason why Shannon's communication model is not ideally suited for studying biological networks relies upon the fact it is solely based on the idea of a statistical correlation between discrete variables and on the so-called subadditivity principle. As a matter of fact, if two discontinuous variables are correlated, subadditivity principle should apply only if all the nodes of the network are associated with these variables. It appears very likely that this condition is not fulfilled for biological networks. This is precisely what is expressed in the statement made previously that the probability spaces are different for a communication process and a biological network. This means that three possible situations can be expected to occur. The joint entropy of the dynamic process is equal to the sum of the individual entropies of its components. Then, this process does not contain any information and its properties can be reduced to those of its components. If, alternatively, the joint entropy of the network is smaller than the sum of individual entropies of the component sub-systems, the network behaves as an integrated entity whose properties cannot be reduced to those of its elements. The information content of the system is then a measure of its degree of integration. Last but not least, if the joint entropy of the network is larger than the sum of the individual entropies of its components, the system is complex and displays emergent effects associated with negative information.
These three possible situations are precisely found to take place with simple enzyme-catalysed reactions occurring under quasi-equilibrium. In these uncomplicated networks, two substrate-binding processes are obviously not correlated, but they may physically interact, that is the binding of a substrate may facilitate or hinder that of the other. Although there is no correlation between discrete variables, there is an information transfer between two processes. Information theory can be extended to express quantitatively this situation, and one can demonstrate that, if the binding of a substrate facilitates that of the other, the overall system is integrated and its information is positive. Alternatively, if the binding of a substrate hinders that of the other, the system is complex and its information is negative.
Thus, in the case of an enzyme reaction whose joint entropy is smaller than the sum of the corresponding individual entropies, the probability of occurrence of the E·AX·B state is smaller than the product of the probabilities of occurrence of E·AX and E·B. This implies in turn that the fundamental energy level of E·AX·B is low, leading to a high-energy barrier between this state and the corresponding transition state of the chemical reaction. If, conversely, the joint entropy is larger than the sum of individual entropies, the probability of occurrence of the ternary E·AX·B enzyme–substrates complex is destabilized relative to the binary E·AX and E·B complexes. In the former case above, one should expect the catalytic rate constant of the reaction to be small, whereas in the latter the catalytic constant should be high.
If the network does not occur under quasi-equilibrium but under steady-state conditions, which implies that it is thermodynamically open, the departure from quasi-equilibrium alone generates emergence of information. This situation may take place even in the absence of any physical interaction between the substrate binding processes. Departure from equilibrium may thus be sufficient to generate complexity. It therefore appears that, within the frame of the definitions given above, apparently uncomplicated biochemical systems may be considered complex.



