1 Introduction
One of the early surprises to emerge from genome sequencing projects was the small number of genes relative to the number of known proteins. Much of the complexity of the proteosome can be attributed to alternative splicing [1]; in the most extreme case of alternative splicing described to date, the Dscam gene alone could potentially encode more than 38,000 different protein isoforms [2]. There are additional sources of diversity within the genome, however, including the use of alternative promoters and polyadenylation sites [3], which can lead to differences at the 5′ and 3′ ends of mRNAs, and RNA editing, which can affect gene expression in a variety of ways. This review briefly summaries the impact of RNA editing on the coding capacity of eukaryotic genomes.
2 RNA editing
2.1 Distribution of RNA editing
The term RNA editing was initially coined by Benne and colleagues to describe the insertion of four non-encoded uridines into the coxII gene of kinetoplastid protozoa [4]. It is now used to describe any specific change in the primary sequence of an RNA molecule, excluding other mechanistically defined processes such as RNA splicing or polyadenylation. RNA alterations due to editing fall into two broad categories, depending on whether the change happens at the base or nucleotide level. The distribution of the best-characterized forms of editing are listed in Table 1 (base substitution) and Table 2 (nucleotide changes). RNA editing is quite widespread, occurring in mammals, viruses, marsupials, plants, flies, frogs, worms, squid, fungi, slime molds, dinoflagellates, kinetoplastid protozoa, and other unicellular eukaryotes (see references in Tables 1 and 2). It should be kept in mind that this list most likely represents only the tip of the iceberg; based on the distribution of homologues of known editing enzymes, for example, editing almost certainly occurs in many other species, including all metazoa [5]. A number of comprehensive reviews on RNA editing are available [6,7], as are recent reviews of base substitution editing [5,8–10] and nucleotide insertion/deletion editing in trypanosomes [11,12].
Editing distribution: base substitutions
| Organism | Editing type | Examples | References |
| Mammals | C to U | apolipoprotein B mRNA | [18,19] |
| A to I | serotonin receptor and | [5,8,49,50] | |
| ion channel mRNAs | |||
| Marsupials | C to U | mitochondrial tRNAs | [51] |
| Plants | C to U | chloroplast mRNAs, | [31,37,38] |
| and | mitochondrial mRNAs, | ||
| U to C | rRNAs, and tRNAs | ||
| Hepatitus delta virus | A to I | HDV antigenome | [52] |
| Drosophila | A to I | ion channels | [53,54] |
| Squid | A to I | ion channels | [55] |
| C. Elegans | A to I | 5′ and 3′ UTRs | [5] |
| Physarum | C to U | coxI mRNA | [33] |
| Trypanosomes | C to U | 7 SL RNA | [22,56] |
| mitochondrial tRNA | |||
| Dinoflagellates | A to G | coxI and cytb mRNAs | [57] |
| G to A | |||
| C to U | |||
| U to C | |||
| G to C | |||
| U to A | |||
| U to G |
Editing distribution: nucleotide insertions and deletions
| Organism | Editing type | Examples | References |
| Kinetoplastids | U insertion | mitochondrial | [4,11,12,29] |
| U deletion | mRNAs | ||
| Physarum | C insertion | mitochondrial | [33,36,58] |
| U insertion | mRNAs | ||
| UU insertion | tRNAs | ||
| AA insertion | rRNAs | ||
| UA insertion | |||
| CU insertion | |||
| GU insertion | |||
| GC insertion | |||
| Paramyxovirus | G insertion | P mRNA | [28,47] |
| Ebola virus | A insertion | GP mRNA | [59,60] |
| Nematodes | U insertion | cytb mRNA | [15] |
| Acanthamoeba | deletion/insertion | mitochondrial | [48] |
| C to A | tRNAs | ||
| A to G | |||
| U to G | |||
| U to A |
2.2 Mechanisms of editing
Editing occurs via a variety of mechanisms, only a few of which have been described in detail [7]. Most characterized instances of base substitutions are due to deamination reactions involving either cytidine (which is converted to uridine) or adenosine (which is converted to inosine) within the context of an RNA molecule. Specificity at these sites can be linked to cis-acting elements within the RNA and the activities that carry out the editing mechanism. In the case of A to I changes in mammalian mRNAs, base-pairing between intron and exon sequences creates a double-stranded region that is recognized by ADARs (
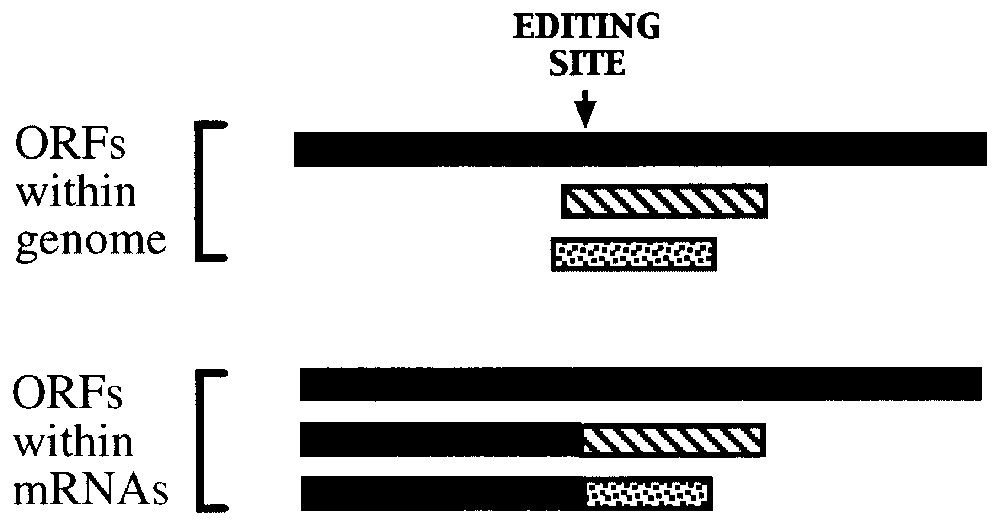
Nucleotide insertion/deletion editing can also occur via a variety of mechanisms. In paramyxoviruses, the viral RNA polymerase ‘slips’ on a homopolymer tract [14]; a similar mechanism may be responsible for the occasional U insertions within the cytb transcript in the nematode Teratocephlus lirellus [15] (Fig. 1). In contrast, although nucleotide insertions into mitochondrial transcripts in Physarum are co-transcriptional, all evidence indicates that it occurs via a distinct mechanism [16]. Finally, U insertion and deletion within kinetoplastid mRNAs is a post-transcriptional process, involving trans-acting guide RNAs (gRNAs) and a number of enzymatic activities [17], as described in Section 3.1.

Nucleotide insertion and deletion patterns. Comparison of short regions of genomic and RNA sequences found in the coxIII gene of Trypanosoma brucei [46], coxI of Physarum polycephalum [33], the P gene of Sendai virus [47], the cytb gene of Teratocephlus lirellus [15], and the tRNAasp gene of Acanthameoba castellanii [48]. Upper case letters indicate nucleotides encoded in the genome, lower case letters indicate nucleotides added by RNA editing.
2.3 Functions of RNA editing
Because editing affects the primary sequence of an RNA, most editing events impact gene expression. Base substitutions most often lead to changes at the amino acid level, whereas the insertion and deletion of nucleotides result in frameshifts in mRNAs (Fig. 2), creating new open reading frames (ORFs). Both types of editing can also affect RNA secondary structure in tRNAs and rRNAs and create (or destroy) start and stop codons. For example, as illustrated in Fig. 3, a single C to U change within the apolipoprotein B mRNA changes a glutamine codon (CAA) to a stop codon (UAA), leading to the production of two proteins from a single gene [18,19]. Other processes that can be affected include RNA splicing, transport, and stability. The editing enzyme ADAR2 edits its own mRNA to create an alternative splice site, providing a potential auto-feedback mechanism [20]. (Other potential links between splicing and editing are discussed in [5,8].) RNAs that contain many inosines are retained in the nucleus [21], and a number of RNAs are known to be edited within 5′ and 3′ untranslated regions (UTRs), potentially affecting stability [5]. Editing of tRNAs can change the ‘identity’ of the tRNA via changes in its anticodon, create substrates for base modification, or create secondary structures essential for processing [10,22]. Not all editing events have obvious effects, however, as some codon changes are silent, while others fall within introns and non-coding regions of mRNAs.
Nucleotide insertion into paramyxoviral RNAs creates multiple open reading frames through frameshifting during RNA synthesis.
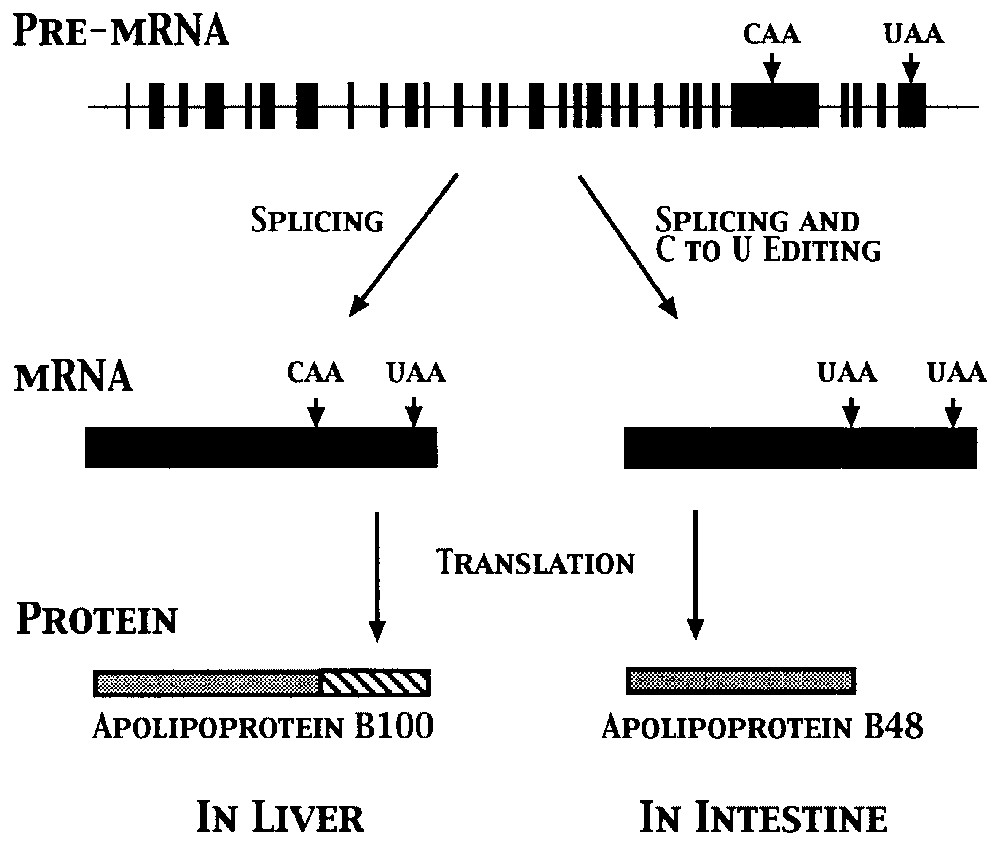
Production of two forms of apolipoprotein B via RNA editing occurs in a tissue-specific manner. Conversion of a single cytidine to a uridine (C to U) results in the creation of an additional stop codon in apoB mRNA in the intestine, leading to the production of a shorter protein.
In many cases, partially edited molecules are also functionally significant. For example, the addition of a variable number of nucleotides at the single editing site within the P mRNAs of paramyxoviruses allows all three reading frames to be accessed in this region of the gene (Figs. 1 and 2). Similarly, partial editing at the 5 A to I sites within the serotonin (5-HT2C) receptor mRNA results in the production of multiple mRNAs; thus far 18 different cDNA sequences and 12 different predicted protein isoforms have been reported (Fig. 4, [23]). Interestingly, the ratio of the individual isoforms varies in different regions of the brain, and at least some have altered G protein coupling properties, suggesting that many of the predicted protein products are likely to be functionally important [24].
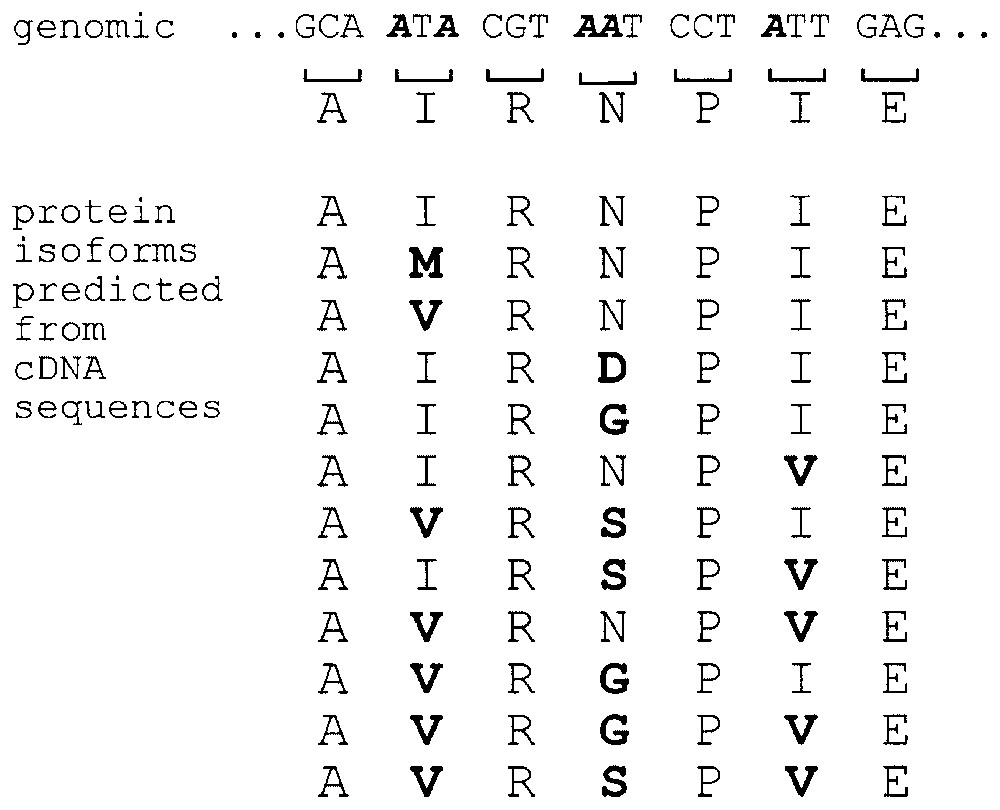
RNA editing leads to the production of multiple isoforms of the serotonin 2C receptor from a single gene. Top: Genomic sequence of a portion of the mRNA encoding the serotonin receptor. The five adenosines that are subject to deamination are in bold italics. The protein sequence predicted from the genomic sequence is directly below each codon. Bottom: Predicted amino acid sequences based on the sequence of known cDNAs. Based on data in [23,49].
The importance of certain editing events has been convincingly demonstrated through gene knockouts of editing enzymes. For example, the gene encoding an RNA ligase required for uridine insertion into kinetoplast mRNAs is essential for survival of the bloodstream form of Trypanosoma brucei [25]. ADAR knockouts in flies and worms lead to behavioral abnormalities, including defects in chemotaxis in worms [5] and locomotion, grooming, and mating in flies [26], while ADARs are absolutely essential in mammals (see Section 3.2).
2.4 Patterns and efficiency of editing
Even when the same types of changes occur in different organisms, editing patterns vary considerably between species. For example, only a single C to U change is observed within the 14,000 nt apoB mRNA in mammalian cells, while the identity of nearly 14% of the encoded residues within the nad3 mRNA in wheat mitochondria are affected by C to U changes [27]. Similarly, changes at the nucleotide level can range from the insertion of a single G, as observed in the measles virus P mRNA [28], to the post-transcriptional addition of more than 50% of the nucleotides within mRNAs initially transcribed from ‘pan-edited cryptogenes’ in kinetoplasts [29] (Fig. 1). Patterns of nucleotide insertion are particularly diverse in regards to the sites of nucleotide insertion and the nucleotides that are added, as can be seen in the examples illustrated in Fig. 1 and Table 2.
In cases where editing is limited to a small number of discrete sites, there is usually a particular sequence that is responsible for directing editing to that site. Examples of this include the ‘mooring sequence’ downstream of the C to U conversion site within the apoB mRNA editing site and the homopolymer tracts found in viral systems. Surprisingly, where editing is more widespread, signals have generally been more difficult to identify. In Physarum mitochondria, for example, no consensus sequence surrounding editing sites has emerged, despite the fact that over 400 C insertion sites have been characterized [30]. Editing contexts are not entirely random in this system, as roughly 70% of the precisely mapped editing sites fall after a purine-U. There is also some codon bias to both base conversions in plant mitochondria [31] and addition of non-templated nucleotides to slime mold mitochondrial mRNAs [30,32], but the basis of these biases is currently unknown.
The efficiency of editing also varies considerably between species. For example, essentially all RNAs present in Physarum mitochondria are fully edited [33], while in kinetoplasts, a significant percentage of the steady-state pool of RNAs is made up of unedited or partially edited molecules [34]. This difference is largely due to differences in the mechanisms used to insert extra nucleotides in these two organisms. Except in cases where start or stop codons are created or destroyed, the efficiency of editing is often less critical in instances of base conversion, as both the edited and unedited forms of the mRNA are likely to produce a protein with at least some function, but editing at the Q/R site within the glutamate receptor B subunit (gluR-B) mRNA is essential in mice [35].
2.5 Regulation of RNA editing
RNA editing is subject to regulation at many levels. Base changes in human cells are tissue specific, with A to I changes occurring primarily in neuronal tissues, while apoB editing occurs only in the intestine. Some of these events are also regulated developmentally, hormonally, or environmentally [8]. Likewise, uridine insertion/deletion in many trypanosome mRNAs is developmentally regulated, occurring in only a single life cycle stage [11]. Expression of editing enzymes is also highly regulated, and multiple isoforms are sometimes produced via alternative splicing [5]. This area of research is likely to expand once more editing targets are identified.
3 Implications of RNA editing
3.1 Implications for gene discovery
The existence of RNA editing complicates gene discovery efforts, particularly in cases where start or stop codons are created (or destroyed) or nucleotides are added or deleted. In Physarum mitochondria, for example, traditional gene finding programs were unable to identify the genes for nad2, nad4L, nad6, and atp8, despite the fact that the entire mitochondrial sequence had been determined and it was suspected that these mRNAs were edited [36]. We are currently collaborating with Dr Ralf Bundschuh on developing specialized programs capable of recognizing such ‘cryptogenes’. His current algorithm, which is based on protein alignments, has recently been used to localize uncharacterized Physarum mitochondrial genes and predict nucleotide insertion sites with high accuracy (J. Gott and R. Bundschuh, unpublished data). Protein alignments also played a key role in the discovery of editing in plant mitochondria [37,38]. More often, however, instances of editing are discovered by accident, through the comparison of genomic and cDNA sequences. Experimental confirmation is essential, particularly given the error rates of EST sequences.
Perhaps the most serious challenge to the concept of the gene is provided by kinetoplastid ‘genes’ in trypanosomes. The kinetoplast, the single mitochondrion at the base of the flagella of trypanosomes, contains a concatenated network of DNA molecules comprised of ∼20–50 maxicircles and ∼5000–10,000 minicircles (Fig. 5) [29]. Pre-edited mRNAs are produced from ‘cryptogenes’ encoded in the maxicircles, which are not functional without editing. The missing information is ‘encoded’ in antisense gRNAs, most of which are transcribed from minicircles [17]. The information in the gRNAs is not translated directly; instead, proteins encoded in the nuclear genome use gRNAs as ‘templates’ to guide the addition or subtraction of uridine residues opposite As or Gs in the guiding region of the gRNA [11]. Thus, three different classes of DNA molecules (maxicircles, minicircles, and the nuclear genome) are needed to produce functional mitochondrial mRNAs that, in most other organisms, are encoded in a traditional manner [12].
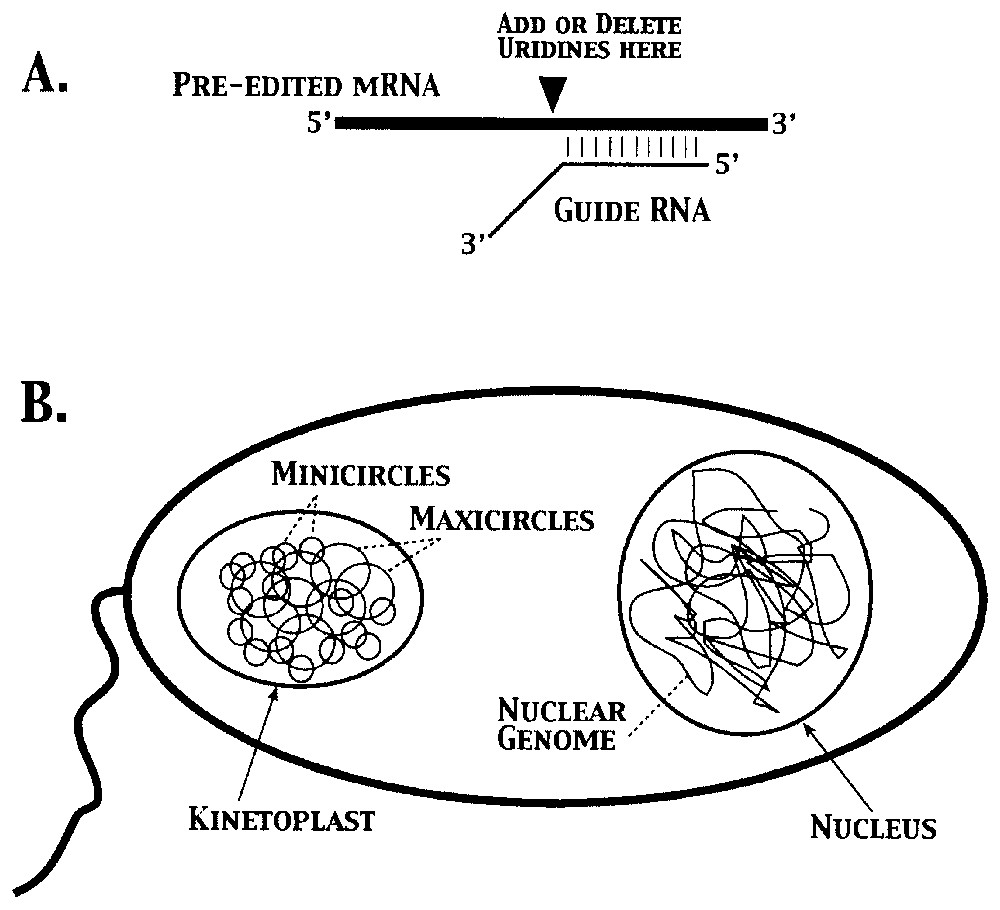
Creation of functional mRNAs in kinetoplasts requires contributions from three classes of DNA molecules. A. Transfer of genetic information from guide RNAs transcribed from minicircle DNAs to pre-edited mRNAs derived from maxicircle DNAs requires enzymatic activities encoded in the nuclear genome. Note that synthesis of a functional mRNA requires incorporation of nucleotides that are the complement of those found in the guide RNAs. B. Schematic representation of a trypanosome, showing the DNA molecules present in the kinetoplast and nuclear compartments (not to scale). Masquer
Creation of functional mRNAs in kinetoplasts requires contributions from three classes of DNA molecules. A. Transfer of genetic information from guide RNAs transcribed from minicircle DNAs to pre-edited mRNAs derived from maxicircle DNAs requires enzymatic activities encoded in the nuclear genome. ... Lire la suite
3.2 Implications for human disease
As with any process that affects gene expression, RNA editing has the potential to go awry. Hyperediting caused by overexpression of Apobec-1 leads to carcinomas in model systems [39], while hyperediting of measles transcripts has been observed in patients with subacute sclerosing panencephalitis and measles inclusion body encephalitis [28,40]. The three ADAR genes are essential in mammalian systems [5]. Deleting even a single ADAR1 allele is embronically lethal in mice; single knockouts have severe defects in the hematopoietic system [41]. ADAR2 knockout mice are prone to seizures and die shortly after birth [35]. Altered editing levels have also been observed in malignant gliomas [42], schizophrenic patients [43] and suicide victims [44], and may be affected in patients with Alzheimer's and Huntington's disease [45]. Finally, editing may also have important implications for drug therapy, since 5-HT2c receptors translated from edited and unedited mRNAs have different affinities for some antipsychotic drugs [44]. Thus, it is clear that RNA editing both expands the coding capacity of the genome and has a significant impact on gene expression.
Acknowledgements
Work in the author's laboratory is supported by a grant from NIH (GM54663).


