1 Introduction
Brown-midrib (bmr) mutants exhibit a reddish brown pigmentation of the leaf midrib and stalk pith, associated with lignified tissues, after the plants have about five expanded leaves. To date, bmr phenotypes have only been seen in maize, sorghum and millet [1], which are all diploid C4 plants belonging to the Panicoideae subfamily in the Poaceae (grass) family (monocotyledons). Plants of the grass family have well-conserved genomes, and likely have a monophyletic origin. According to recent phylogeny data [2,3], most grasses comprise two clades. The oldest diverging clade (BEP), in which no bmr mutants were found, is comprised of Bambusoideae, Ehrhartoideae (about 105 species, including rice) and Pooideae (about 3300 species, including cereal crops and temperate C3 forage grasses). The earlier diverging clade (PACCAD) is comprised of Panicoideae (about 3250 species), and five others subfamilies (Arundinoideae, Centhothecoideae, Chloridoideae, Aristidoideae, and Danthonioideae). Most Panicoideae are characterized by the C4 Krantz photosynthesis, and have only one layer of bundle sheath cells surrounding their vascular bundles. However, no relationships are clearly established between the C4 anatomy and the occurrence of bmr mutants.
The first bmr maize plant appeared in a self-pollinated line of a northwestern dent maize in 1924 [4]. The gene was subsequently named bm1 and three other genes inducing the bmr phenotype were described later, as bm2 in 1932 [5], bm3 in 1935 [6], and bm4 in 1947 [7]. Each bmr gene of maize originates from natural mutations and segregates as a simple Mendelian recessive character. The effects of maize bmr mutations on lignin content and forage feeding value were evidenced latter [8,9]. In sorghum, 19 independently-occurring bmr mutants were identified in segregating progenies of chemically treated seeds of two lines [10]. Some of these mutants had significantly reduced lignin content, and/or a significantly higher cell-wall digestibility. Bmr mutants in pearl millet also originated from chemically-induced mutations [11].
Many studies were then made on bmr plants, used as models in digestibility and lignification studies (reviewed in [1,12,13]). Studies of lignin content and structure, and p-hydroxycinnamates in the cell walls of the maize bm3 mutant, which is the one with the higher efficiency for improving plant stover digestibility, provided one of the first significant clues to the means by which lignification impacted digestibility. In addition to the bm3 maize model, other maize mutants together with the bmr mutants of sorghum and millets have already provided, and will probably further provide, important information on the monolignol biosynthetic pathway and the most promising targets for cell-wall digestibility improvement. The objective of this text is to give an overview of available biochemical and molecular data on bmr genotypes, and then to assess the more likely pathways for lignin and hydroxycinnamic acid biosynthesis in grasses.
2 Genetic variation in lignin structure and cell wall cross linking in maize brown-midrib plants
2.1 The bm3 mutant of maize
The bm3 mutant is the one on which most investigations have focused, probably because it is the most efficacious in feeding value improvement. A quasi-lack of caffeic acid O-methyl transferase (COMT) activity was established in maize bm3 plants [14], corresponding to an altered exon 2 of this COMT [15,16]. In the sixties, Kuc and Nelson [8] established that the frequency of syringyl (S) units was heavily reduced, and suspected the occurrence of additional not yet detected units. These additional units were identified latter as 5-hydroxyguaiacyl (5-OH-G) units [17]. In bm3 maize and in COMT-deficient plants, these 5-OH-G units were shown to be involved in novel benzodioxane structures in the lignins [18–21]. Bm3 mutants had a lignin content reduced by 25 to 40%, but relative to their lignin contents, bm3 plants may be less digestible than expected. Lignins of bm3 appear to inhibit proportionally more cell-wall digestibility than normal lignin [22], which could be related to a higher frequency of coniferaldehyde units in the polymer [23].
Later research on COMT specificity and on lignins in various COMT-deficient dicotyledonous plants demonstrated that COMT is primarily involved in the formation of S lignin units via methylation of 5-hydroxyconiferaldehyde [24–27], and could then be considered as a 5-hydroxyconiferaldehyde O-methyltransferase (‘CaldOMT’). However, even when this COMT is completely deficient in bm3 maize, S lignin units are heavily depleted, but not absent. S units may therefore originate from another non-described pathway, or 5-hydroxyconiferaldehyde might be a possible substrate of caffeoyl Coenzyme A O-methyltransferase (CCoAOMT), despite the statement that “there is as yet no report of the activity of this enzyme towards the aldehyde or alcohols” [28].
In agreement with pioneering and recent literature data [8,18,29–31], results given in Tables 1 and 2 further confirmed that bm3 stems collected at the silage stage have less lignin, and a lower frequency of S lignin units. They also have fewer p-coumarate esters, a reduction consistent with the preferential acylation of S units by p-coumaric acid (pCA) [32–34]. These studies further confirmed that bm3 plants were not altered in their content of alkali-releasable ferulic acid (FA). Similar data have been recently obtained with a maize COMT antisense line [35]. In younger plants, Marita et al. [18] obtained a greater amount of FA esterified to arabinoxylans in the bm3 cell walls, but similar cross-linking between arabinoxylans (total FA dimers). These results strengthened the hypothesis that the bm3 gene is not involved in FA biosynthesis. Relative to the control line, the bmr18 mutant of sorghum, which had a lower OMT activity, displayed similar ester and ether FA contents, but less pCA esters [36,37]. Recently, premature stop codons were shown in the COMT gene of bmr12 and bmr18 sorghum mutants [38].
Comparative analysis of stems in regular and brown-midrib isogenic lines [IVNDFD=in vitro neutral detergent fiber digestibility (weight%); KL/NDF=Klason lignin content (weight% NDF); ester p-CA, ester FA, ether FA=contents in esterified p-coumaric, ferulic acids, and etherified ferulic acid, respectively (mg g−1 NDF)]. Mean values and standard errors correspond to independent experiments run on the three replicates of field trial for each line
| NDF | IV NDFD | LK/NDF | ester pCA | ester FA | ether FA | |
| F292 | 56.2 | 15.9 | 16.4 | 49.5 | 11.9 | 3.2 |
| F292bm1 | 54.9 | 19.2 | 14.6 | 26.0 | 9.8 | 2.4 |
| F292bm2 | 51.3 | 20.2 | 13.6 | 45.5 | 12.4 | 1.8 |
| F292bm3 | 52.2 | 23.1 | 13.0 | 28.5 | 12.8 | 3.0 |
| F292bm4 | 53.4 | 22.8 | 14.3 | 45.1 | 12.6 | 2.6 |
| F2 | 49.2 | 20.1 | 18.0 | 40.0 | 10.3 | 4.6 |
| F2bm1 | 46.7 | 25.7 | 14.0 | 21.8 | 10.6 | 2.3 |
| F2bm2 | 45.7 | 20.5 | 15.2 | 35.9 | 13.6 | 1.7 |
| F2bm3 | 46.0 | 28.4 | 13.3 | 20.9 | 12.3 | 4.7 |
| Standard error | 1.2 | 2.2 | 0.5 | 0.9 | 0.3 | 0.8 |
Analysis of the main lignin-derived monomers obtained by thioacidolysis of extract-free stems from stems in regular (F2, F292) and brown midrib (F2bm1, F2bm2, F2bm3, F292bm1, F292bm2, F292bm3, and F292bm4) lines (total yield of p-hydroxyphenyl H, guaiacyl G, syringyl S, and 5-hydroxyguaiacyl 5-OH-G monomers expressed in μmoles per gram of Klason lignin. Mean values and standard errors correspond to independent experiments run on the three replicates of field trial for each line)
| H | G | S | 5-OH-G | S + G | S/G | |
| F292 | 18.4 ± 0.7 | 325 ± 12 | 547 ± 25 | 3.0 ± 0.06 | 872 | 1.68 |
| F292bm1 | 11.7 ± 0.4 | 182 ± 18 | 286 ± 31 | 2.0 ± 0.06 | 468 | 1.57 |
| F292bm2 | 18.5 ± 0.7 | 298 ± 14 | 557 ± 42 | 2.8 ± 0.05 | 855 | 1.87 |
| F292bm3 | 9.7 ± 0.4 | 436 ± 20 | 231 ± 16 | 74.0 ± 5 | 667 | 0.53 |
| F292bm4 | 15.0 ± 0.5 | 264 ± 27 | 514 ± 48 | 3.1 ± 0.3 | 778 | 1.95 |
| F2 | 12.7 ± 0.6 | 442 ± 22 | 453 ± 22 | 1.2 ± 0.04 | 895 | 1.02 |
| F2bm1 | 6.6 ± 0.3 | 225 ± 18 | 227 ± 16 | 1.1 ± 0.06 | 452 | 1.01 |
| F2bm2 | 12.5 ± 0.6 | 286 ± 4 | 400 ± 32 | 2.6 ± 0.05 | 686 | 1.40 |
| F2bm3 | 7.8 ± 0.3 | 531 ± 31 | 183 ± 17 | 71 ± 6 | 714 | 0.34 |
2.2 Comparisons between maize brown-midrib mutants
Although many data are now published on the structure of lignins in bmr maize lines, no systematic and recent comparison of the four bmr genes in an isogenic genetic background in maize harvested at the silage stage seems to be available. Original data for a series of bm1, bm2, bm3, and bm4 mature mutant lines obtained in the same genetic background are available from an unpublished investigation at the INRA Lusignan and Grignon centers.
The maize bm1 mutation specifically affects the expression of a cinnamyl alcohol dehydrogenase (CAD) gene, the activity of which was reduced by 60–70% in the above-ground plant and by 90–97% in roots [39,40]. Maize bm1 stems had reduced lignin content, a slight decrease in FA esters, and a substantially reduced content of pCA esters and FA ethers (Table 1). Similar trends were observed in younger bm1 plants at the emerging tassel stage [18], except that the lignin content was unmodified. The reduced recovery of thioacidolysis monomers (Table 2) reveals that the frequency of lignin units involved only in β-O-4 bonds was about 50% lower in bm1 than in the control. Bm1 lignins appeared to be substantially enriched in carbon–carbon inter-unit linkages. The frequency of p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) thioacidolysis monomers was similar in the bm1 and control samples, showing that the bm1 mutation does not specifically affect a lignin unit type, similarly to CAD down-regulated tall fescue [41], but contrarily to CAD-deficient poplars [42,43]. Specific thioacidolysis markers, diagnostic for the incorporation of coniferaldehyde and/or sinapaldehyde units into the lignins, were investigated (Table 3 and Fig. 1) [42,43]. Coniferaldehyde end-groups linked to lignins at their phenolic oxygen give rise to a specific dithioketal derivative (Fig. 1A). The S analogue was only recovered as a trace component, which means that sinapaldehyde is not prone to be incorporated into lignins as end-groups. When coniferaldehyde or sinapaldehyde units are linked at their Cβ-carbons (Fig. 1B), they give rise to specific bicyclic isomers [42]. The data in Table 3 give the first direct evidence for the substantial incorporation of coniferaldehyde and, to a lower extent, of sinapaldehyde into native bm1 lignins. While the p-OH cinnamaldehyde-derived compounds were recovered in low or trace amounts from the regular, bm2, bm3, and bm4 lines, their relative importance was found considerably (50 to 150 fold) increased in the bm1 lignins. The amount of coniferaldehyde linked at C4OH or at Cβ exceeds that of sinapaldehyde linked at Cβ (Table 3). Lignins of bm1 maize mutants thus incorporate both coniferaldehyde and sinapaldehyde as a result of CAD deficiency, a phenomenon previously reported for the CAD-deficient bmr6 sorghum mutant [44]. In contrast, sinapaldehyde was found to be specifically incorporated into the lignins of CAD-deficient poplar [20,42,43], Arabidopsis [45], while the coniferaldehyde level was not affected. According to NMR data, both coniferaldehyde and sinapaldehyde were incorporated into lignins of CAD-down-regulated tobacco [20,46]. As studies on synthetic lignins made from coniferaldehyde [44,47] and on CAD-deficient pine [47,48] have shown that coniferaldehyde incorporates into lignins not only through labile ether bonds, but also through resistant biphenyl structures, the total amount of coniferaldehyde in bm1 lignins probably exceeds the amount of coniferaldehyde-derived thioacidolysis monomers. Thioacidolysis of bm1 native lignins also give rise to the dithioketals of vanillin and syringaldehyde in similar amounts as the p-OH-cinnamaldehyde-derived compounds (data not shown). This result suggests that p-OH-benzaldehydes incorporate into bm1 lignins and/or that p-OH-cinnamaldehyde lignin units are easily degraded to vanillin and syringaldehyde [47].
Thioacidolysis signatures for the incorporation of coniferaldehyde and sinapaldehyde units in the lignins of regular and bmr maize lines: yields (expressed in μmole per gram of Klason lignin). 1: Marker for the incorporation of coniferaldehyde end-groups (structure A in Fig. 1); 2: marker for the incorporation of coniferaldehyde units linked at their Cβ carbon through β-O-4 bonds (structure B in Fig. 1, with R=H); 3: marker for the incorporation of sinapaldehyde units linked at their Cβ carbon through β-O-4 bonds (structure B in Fig. 1, with R=OMe); tr=trace amount. Standard errors between triplicates are in the 5–10% range
| Line | 1 from coniferaldehyde end-groups | 2 from coniferaldehyde linked at Cβ | 3 from sinapaldehyde linked at Cβ |
| F292 | 0.9 | tr | 0.7 |
| F292bm1 | 8.6 | 26.8 | 14 |
| F292bm2 | 0.8 | tr | 0.7 |
| F292bm3 | 1.7 | tr | tr |
| F292bm4 | 0.8 | tr | 0.6 |
| F2 | 1.9 | tr | 0.5 |
| F2bm1 | 5.9 | 18.7 | 12 |
| F2bm2 | 0.7 | tr | 0.4 |
| F2bm3 | 2.2 | tr | tr |
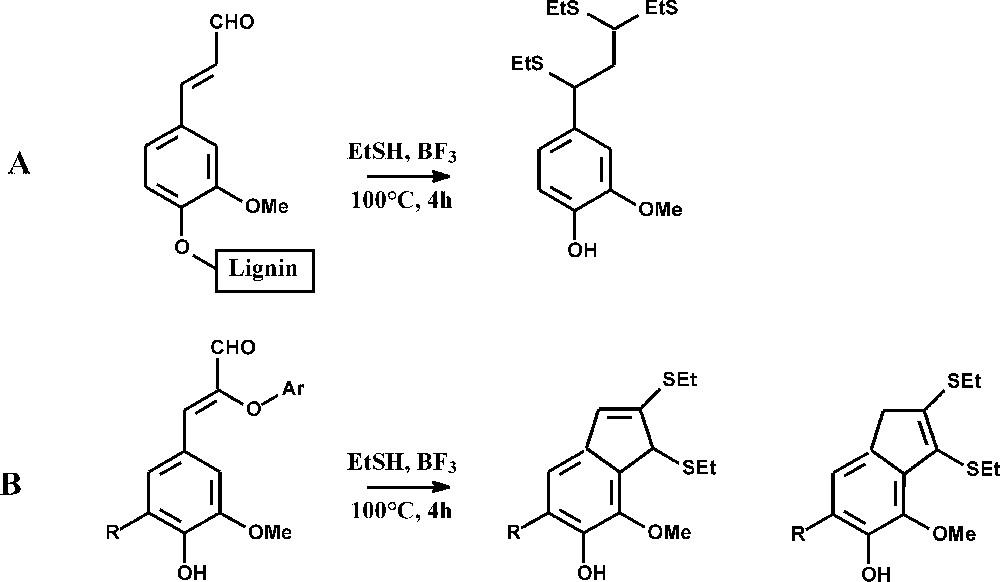
Thioacidolysis of coniferaldehyde end-groups (A) and of coniferaldehyde (R=H) or sinapaldehyde (R=OMe) units linked to their Cβ carbon through β-O-4 bonds.
In the case of aspen, Li et al. [49] established the occurrence of two alcohol dehydrogenase activities, corresponding to PtCAD and PtSAD cDNA, and respectively associated with the formation of G and S lignin units. In Arabidopsis, null mutants Atcad-C and Atcad-D (similar to CAD4 and CAD5 [50]) had a drastically reduced CAD activity, with a higher impact on SAD activity [45]. The precise target of the bm1 mutation (CAD and/or SAD) is also unclear. To date, only one CAD gene involved in constitutive lignification was evidenced in maize (bm1, in bin 5.04), but other CAD or CAD-like genes likely exist, such as the CAD2 gene in bin 2.03 [51]. Three CADs were sequenced in a C3 grass, Lolium perenne [52], the LpCAD3 gene being the closest to the maize CAD gene, and four close genes were evidenced in a cDNA library of Festuca arundinacea [41]. Moreover, in Eucalyptus gunnii, the occurrence of two functionally and structurally distinct CAD proteins, CAD-1 and CAD-2, was established [53,54]. The latter was speculatively suggested as involved more in constitutive lignification. In several plant species, CAD-2 type proteins have been found to be encoded by a single gene, or by a small multi-gene family [54,55]. In maize, CAD-2 like enzyme(s) involved in the lignin pathway are probably dimeric forms, with different affinities according to their homodimer and heterodimer status, as it was observed in Eucalyptus gunnii [56]. Conversely, CAD-1 was shown to be monomeric in Eucalyptus gunnii, unable to use sinapaldehyde as substrate, and thereby considered to function as an ‘alternative pathway’ in lignin biosynthesis [54,57]. In vitro NDF digestibility (IVNDFD) of bm1 lines was (slightly) higher than in regular lines, but lower than in isogenic bm3 lines (Table 1, [58]). In contrast, CAD down-regulated plants displayed substantially improved performance with regard to chemical pulping due to the alteration in lignin structure [59].
A reduced lignin content, a slight shift in pCA and FA ester levels, and a significant decrease in FA ether levels were observed in mature bm2 plants (Table 1). The reduced level in FA ethers may result from FA esters incorporating into lignins through other linkage modes, as evidenced in the case of normal plants [60]. Conversely, on younger plants, an elevated level of etherified FA was observed in bm2 plants [18]. In agreement with previous data [61], the frequency of β-O-4-linked G units was substantially reduced in bm2 plants, whereas the frequency of β-O-4-linked S units was not affected. A similar trend was observed in F292bm4. Thioacidolysis of bm2 and bm4 lignins did not reveal any incorporation of unusual units, in contrast to bm3 and bm1 lignins. No information is currently available on the genes affected in the bm2 or bm4 mutations. Today, it remains difficult to assign any enzymes directly involved in monolignol biosynthesis to either bm2 or bm4 mutations. Vermerris and Boon [62] hypothesized that the bm2 mutation could affect (i) a transcription factor involved in the spatio-temporal deposition of lignin, (ii) a protein involved in the stereospecific linkages of G units or (iii) coniferyl alcohol (or its glucoside). It remains also possible that bm2 and bm4 mutations affect others steps such as the transportation of monolignols to the walls.
A current dilemma is why bmr plants acquire their brown midribs, and correlatively, why bmr plants are seemingly observed only among (diploid) Panicoideae C4 grasses. To date, no bmr plants have been reported in diploid rice lines. However, different brownish or red colorations were observed in the xylem of CAD-deficient transgenic dicotyledons and in the CAD mutant of loblolly pine [63–65], a situation reminiscent of the wine-red coloration of coniferaldehyde-derived in synthetic lignins [66]. Conversely, no coloration was observed in a COMT mutant nor in CAD downregulated Arabidopsis [45,67,68], but COMT and CAD appear to belong to multigenic families in Arabidopsis [50,68]. The reason for the brown midrib in mutant grasses is therefore not definitively resolved.
3 Questions and hypotheses regarding OMT genes involved in the grass lignin pathway
Until recently, the biosynthetic pathway leading to lignins and p-hydroxycinnamic acids was considered as a metabolic grid, through which side-chain reductions, ring hydroxylation and methylation could occur at different levels. Such a model has recently been markedly reassessed (in dicotyledons plants) by evidence arising from enzymatic substrate preferences, spatio-temporal localization of monolignol biosynthesis enzymes, gene sequencing and transgenic approaches. Different documented reviews were then published illustrating the ‘lignin roadmap’ [26,27,69–72]. According to Dixon et al. [26], two hypothetical independent channels would lead to G and S units. The G pathway would involve hydroxylation of a coumarate moiety, likely at the level of conversion of coumaroyl-CoA to caffeoyl-CoA, followed by its methylation by a CCoAOMT, and reduction of feruloyl-CoA to coniferyl alcohol via coniferaldehyde. The S pathway would involve the methylation of caffeyl aldehyde by a COMT, the conversion of coniferaldehyde to 5-hydroxyconiferaldehyde by a F5H, its methylation by a COMT (the same or a different COMT as for caffeyl aldehyde), and a final reduction to sinapyl alcohol. However, Zhong et al. [73] established that down-regulating CCoAOMT reduced both G and S lignins units, indicating that the CCoAOMT is essential both for the methylation of the 3-hydroxyl group of caffeoyl-CoA and to channel substrates for 5-methylation of 5-hydroxyconiferaldehyde.
The reduction in lignin content of bm3 plants did not affect the yield of G thioacidolysis monomers (Tables 1 and 2). This result reinforces the alternative model of Dixon et al. [26] for independent pathways to G and S lignin units, with a CCoAOMT-catalyzed methylation of caffeoyl-CoA in the G pathway. No caffeyl moiety could be evidenced in bm3 lignins [17,44], whereas the 5-OH-G units are now well established in a variety of COMT-deficient plants [17–21,74]. The CaldOMT disrupted in the bm3 mutation is thus not involved in caffeyl aldehyde methylation. The existence of another COMT specific of caffeyl moieties is unlikely, at least in maize because a correlative ‘bm5’ mutant should have been found. Based on all these data, CCoAOMT is probably a major hub in controlling the flux to both G and S units (Fig. 2). Because at least two CCoAOMTs exist in maize, it could be assumed that either two different genes are involved in G and S unit biosynthesis, or that two (or more) independent mechanisms and transcription factors regulate the two routes to G and S units based on the same CCoAOMT gene. Data are not yet available concerning the respective roles or tissue specificities of the (different) CCoAOMT gene(s) in maize and other grasses. In poplar, each of the two CCoAOMT genes exhibited precise tissue and cell-specific expression patterns [75]. Moreover, there are still no definitive observations to assert that the routes to G and S units are similar in mono- and dicotyledons, or in C4 (or PACCAD) and C3 (or BEP) grasses, even though grasses are monophyletic.
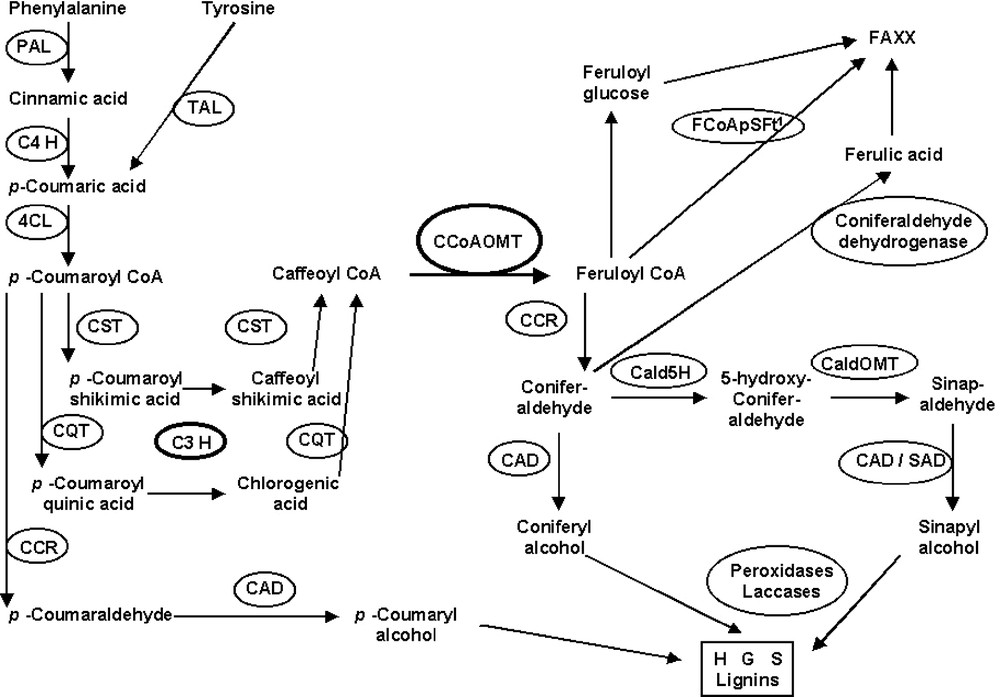
An hypothetical view of the pathways leading to lignins and to ferulolylated arabinoxylan (FAXX) in grasses based on CCoAOMT hub(s) (1=feruloyl-CoA:polysaccharide feruloyltransferase, FCoAPSFt).
The hydroxylation and methylation steps to S ring might also take place at the alcohol level, utilizing the same enzyme complex, but a CAD activity at an earlier stage such as caffeyl aldehyde. From studies in dicotyledons plants, F5H appears to be a coniferaldehyde-5-hydroxylase (Cald5H), and it is therefore logical that hydroxylation and methylation leading to S ring occurs at the aldehyde rather than at the alcohol level [70,76,77]. Osakabe et al. [76] also established that coniferaldehyde inhibited the 5-hydroxylation of ferulate or coniferyl alcohol. However, hydroxylation and methylation steps operating at the alcohol level have been clearly established in magnolia and eucalyptus [78,79]. Chen et al. [78,79] showed that fed coniferin resulted in S units, by di-deutero labeling coniferin at the γ-position, and observing γ-di-deutero-sinapyl acetate DFRC monomers. At least a large fraction of the conversion occurred directly at the alcohol level and not via reversion to the aldehyde, where one of the γ-deuterons would have been lost. The above results also strengthen the notion that, in maize, the bm3 locus encodes a 5-hydroxyconiferaldehyde O-methyltransferase (CaldOMT) rather than a caffeic acid O-methyltransferase (COMT) enzyme (Fig. 2), even though caffeyl aldehyde and 5-hydroxyconiferyl alcohol cannot be excluded as alternative substrates. In C3 grasses, Chen et al. [80] also established that crude extracts of wheat and tall fescue exhibiting O-methyltransferase activities catalyzed in vitro methylation of hydroxycinnamaldehydes and hydroxycinnamyl alcohols more effectively than the corresponding hydroxycinnamic acids. Similarly, in tall fescue, no COMT activity was observed on caffeic acid, when incubating substrates with extracted enzymes from plant tissues [81]. Moreover, the COMT activity was significantly lower, indeed even missing, in the earliest stage of development, which corresponds to synthesis of mostly G lignin units. In contradiction, Lam et al. [82] found in wheat stems an ‘early’ and a ‘late’ OMT activity more efficient on caffeic acid and 5-OH-ferulic acid, respectively. They suggested that the early activity was related to the synthesis of ferulic acid destined for esterification of arabinoxylans, while the late one would be involved in the formation of sinapyl alcohol. However, this interpretation is uncertain as all available results suggest that neither caffeic, nor 5-OH-ferulic, acids seem to be involved in ferulate or sinapyl alcohol biosynthesis in vivo.
Three COMT genes (COMT1, 2, 3) were described in Lolium perenne (2n) [83–86], but one of them (COMT2) is probably not a COMT involved in the lignin pathway [87]. Four COMT genes were described in Festuca arundinacea (6n) [88]. Only one COMT sequence is yet described in databanks for maize, sorghum and, seemingly, sugarcane. Based on the maize genomic COMT (CaldOMT) sequence, only one COMT sequence is found in rice from the Gramene database (http://www.gramene.org). This sequence is located on the short arm of chromosome 8, in position 4,396,266 bp, is named ‘bm3’, and has an 89% identity with the maize sequence. Stinard [89] has located the maize bm3 gene between su1 and gl7 genes, on the short arm 4S, in bin 4.05. Comparisons between maize and rice (Gramene and [90]) showed that a short, reorganized part of rice chromosome 8S occurs in the maize bin 4.05. From the available genomic data, it appears unlikely that the ‘bm3’ rice locus is present elsewhere in the maize genome. Even if molecular or sequence data do not allow definitive conclusions, a unique CaldOMT in maize (and in rice) seems so far the most likely hypothesis. Similarly, from Gramene data, and based on the CCoAOMT1 maize sequence, only one CCoAOMT gene is found in the rice genome, located on rice chromosome 6S, in good agreement with the presence of rice chromosome 6S parts in maize chromosome 6S (maize CCoAOMT1) and 9S (maize CCoAOMT2) [90,91]. Flint-Garcia et al. [51] also located a CCoAOMT3 gene in bin 4.06, which is a heavily reorganized area in the maize genome likely including fragments of rice chromosome 6.
4 Questions and hypotheses regarding genes involved in the ferulic acid pathway, and cross-linking in grasses
Results obtained with the bm3 mutant suggest that ferulic acid is not biosynthesized by the COMT/CaldOMT-catalyzed methylation of a caffeic precursor, since the COMT gene is disrupted in the bm3 plant and yet FA is evidenced. Complicating the issue is that the lower lignin level in the bm3 mutant may increase the yield of alkali-releasable FA, as suggested by the artificial lignification of maize cell walls [92]. The path of ferulic acid biosynthesis still remains questionable in maize, panicoids, and likely in all grasses. According to Fry et al. [93], whereas the cinnamate-to-p-coumarate conversion may occur at the free-acid level, the p-coumarate-to-caffeate and -ferulate conversion could involve conjugates of the acids, or possibly CoA thioesters. The attachment of a feruloyl group to an arabinoxylan might occur by a transacylation involving a feruloyl-CoA:polysaccharide feruloyltransferase. Such a feruloyl-CoA-arabinoxylan-trisaccharide O-hydroxycinnamoyl transferase activity was evidenced in suspension-cultured rice cells [94]. Obel et al. [95] added a new level of complexity, establishing that feruloyl-glucose was the precursor for intracellular feruloylation of arabinoxylan in suspension cultured wheat cells, whereas feruloyl-CoA was the precursor for protein feruloylation. Feruloyl-CoA might also be a donor of ferulic acid in the cell, which could end up as feruloyl-glucose for arabinoxylan feruloylation. Whether either feruloyl-CoA or feruloyl glucose is involved, the ferulic moiety originates from the hydroxycinnamoyl-CoA channel, reinforcing the central role of CCoAOMT in the lignin pathway leading to the lignins and the feruloylated arabinoxylans of grass cell walls (Fig. 2). The resulting ferulic glycosides are then exported to the maturing wall. In the secondary wall, the cross-linking of feruloyl arabinoxylan further involves an active oxidative coupling mechanism, e.g. via peroxidases/H2O2 and/or laccases/O2. Another intriguing pathway for ferulic acid biosynthesis needs to be considered from a very recent result obtained in Arabidopsis. Nair et al. [96] demonstrated that the ref1 mutant, which has a reduced content in soluble sinapate esters, was affected in a sinapaldehyde dehydrogenase. When it was expressed in E. coli, the REF1 gene exhibited both sinapaldehyde and coniferaldehyde dehydrogenase activities. The implication is that the acids in the wall are products beyond the monolignol pathway and derived from oxidation of the corresponding aldehyde, rather than acting as precursors of those aldehydes (and hence the monolignols). Whether this pathway exists in grasses or is limited to Brassiceae (or to dicotyledons plants) is unknown. The existence of a coniferaldehyde dehydrogenase involved in ferulic acid biosynthesis in grasses has thus to be considered as a possible route downstream from C3H and CCoAOMT activities, especially as Arabidopsis REF1 gene shares high amino acid similarity with cytosolic ZmRF2C and ZmRF2D genes from maize or OsALDH1a gene from rice [96].
As a further complicating possibility to be considered for grasses, Schoch et al. [97] have established in Arabidopsis the participation of novel intermediates, such as caffeoyl esters of shikimic and chlorogenic acids, in the 3-hydroxylation of p-coumarate derivatives. Hydroxylation at ring C-3, which is catalyzed by the P450 3-hydroxylase CYP98A3, involved neither free p-coumaric acid nor its CoA ester as previously described, but its 5-O-shikimate and 5-O-D-quinate esters. CYP98A3 was not active toward p-coumaroyl-CoA [97,98]. The ref8 mutant of Arabidopsis, whose lignin content is severely decreased and with lignins essentially composed of H units, instead of G and S units as found in the wild type, is a CYP98A3 mutant [99]. This result indirectly established the lack of another substitutive C3H enzyme in Arabidopsis. Humphreys and Chapple [70] noted that, despite numerous attempts, C3H hydroxylase was one of the last classic phenylpropanoid enzymes and genes remaining uncharacterized.
All these convergent results make the biosynthesis of ferulic acid from caffeic and p-coumaric acids unlikely, and confirm the lack of involvement of COMT at the acid level in ferulic acid biosynthesis. These views are also in agreement with the fact that caffeic acid is not an hydroxycinnamic acid constituent of the wall (like p-coumaric and ferulic acids), as might be expected if ferulic acid was synthesized at the acid level. The implication is that caffeic acid is not synthesized in the lignin pathway (at least not significantly). A second consequence would be that 4CL would have p-coumaric acid as an only substrate (but different 4CLs could be involved in grasses).
Based on the current knowledge of the lignin pathway and on the present discussion, CCoAOMT(s) is (are) probably a major hub in controlling grass cell-wall lignification (and digestibility). CCoAOMTs have a strict affinity for CoA esters, more specifically for caffeoyl-CoA, and no affinity toward corresponding acids. CCoAOMT should be a ‘primitive’ enzyme in relation to COMT (or more definitively to CaldOMT), which should be a more evolved form [100,101]. This hypothesis was strengthened by the fact that only a CoA-OMT was found in the genome of cyanobacterium [101]. Moreover, a CCoAOMT gene, with a high catalytic efficiency for caffeoyl-CoA, exists in a gymnosperm (loblolly pine) [102]. A COMT related enzyme, called AEOMT, having a similar activity on acids and esters-CoA, has also been previously found [103]. An older evolutionary origin for CCoAOMT would also be in agreement with the fact that S units, and FA in grasses, had a CCoAOMT origin, and that no caffeic acid OMT (COMT), but only a 5-hydroxyconiferaldehyde OMT (CaldOMT), exists in vivo in the grass lignin pathway. Besides CCoAOMT, C3H, which is likely the sole 3-hydroxylase in the lignin pathway, is also “a major control point in the production of C3- and C5-substitued phenylpropanoids” [27], as it supplies CCoAOMT with caffeoyl-CoA derivatives. A C3H-CCoAOMT channel could then be considered as a central hub in controlling cell-wall lignification.
Studies of intraspecific variation for FA content in maize also provided new information and allowed some hypotheses to be suggested. In a set of maize lines [104], there was no correlation between esterified FA and etherified FA. In addition, line F286 displayed both a high content in FA esters, and a reduced level in FA ethers. As studies of artificially lignified maize walls suggest that the solvolytic release of FA depends on the degree of cell wall feruloylation and lignification [105,106], the lack of correlation between FA esters and ethers probably occurs because lignin and FA deposition is not uniform among tissues or even within a cell wall [107,108]. In the grass cell wall, polysaccharide ferulate esters undergo radical coupling reactions involving peroxidases and laccases. Ranocha et al. [109] showed that laccase down-regulation caused the accumulation of total soluble phenolics in poplar, and alteration of the cell-wall structure, without lignin content modification. F286 could be altered in a gene or a regulation gene involved in the oxidative binding of FA esters. However, since only ferulate 4-O-ethers are quantifiable by alkaline hydrolysis [60,110], a greater propensity for coupling with more resistant linkages could not be completely excluded in F286, but this last hypothesis appears very unlikely, especially as this line was highly digestible. Another mechanism for FA incorporation into cell walls is probably altered in line F7012, which had a FA ester and total FA content lower than all the other lines, but whose FA ether content was in the variation range of other genotypes. This could be related to different defects in gene(s) involved in FA biosynthesis, coupling with carbohydrates, or involved in glucoside transport. However, variation in ferulate content in F286 and F7012 could also be related to lignin distribution and tissue patterning because ferulate is deposited throughout primary and secondary wall formation in most if not all grass tissues and its release by saponification declines dramatically as cell walls undergo lignification [111].
5 Conclusions
Genetic and biochemical studies on maize (and allied species) bmr mutants, together with work on plants genetically modified in the lignin pathway, allow significant advances in the knowledge of the lignin pathway and hydroxycinnamic acid biosynthesis (Fig. 2). The synthesis developed in this text attempts to hypothesize a lignin and hydroxycinnamic acid roadmap (mostly for grasses or monocotyledons) based on genetic, genomic and biochemical data. Maize may be considered as the leading grass model plant for investigations on lignification and its relationships with cell-wall digestibility. Targets considered of interest in maize could be extrapolated to other grass forages, including C3 plants. C3H and CCoAOMTs are probably major hubs in controlling the biosynthesis of both G and S lignin units and of ferulic acid involved in cross-linkages. The lignin pathway probably resembles a linear channel, with diverging canals, rather than a metabolic grid. But the complex spatio-temporal regulation of genes will also demand a comprehensive examination of the regulation mechanisms. In order to confirm the developed hypotheses, the following questions should be elucidated: (i) whether the C-3 hydroxylation at the aromatic ring effectively occurs via shikimate or quinate derivatives in maize, and thus likely in all grasses, (ii) whether specific and different CCoAOMTs are or are not involved in G and S lignin pathways, and in the arabinoxylan feruloylation pathway, and (iii) whether feruloyl-glucose, feruloyl-CoA or coniferaldehyde is the last precursor in arabinoxylan feruloylation in C4 and in C3 grasses, but also (iv) the respective role of the different COMT genes evidenced in different Pooideae genomes, and (v) the role of peroxidases and/or laccases involved in an active oxidative coupling mechanisms for the cross-linking of feruloyl arabinoxylans in the cell wall. In order to understand grass lignification and phenolic pathways, and correlatively grass cell-wall digestibility, the first steps have been to elucidate structural features of lignins, identify cell-wall cross-linking mechanisms, and identify which genes are involved in the monolignol biosynthetic pathway. It remains a challenge to understand how these genes work together and how these diverse mechanisms impact plant cell-wall digestibility.


