Abbreviations
PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; r-ER, rough-endoplasmic reticulum; KLH, keyhole limpet hemocyanin.
1 Introduction
A highly characteristic feature of woody plants is a hard stem, due to the thick secondary cell walls composed mainly of cellulose, hemicelluloses, and lignin. Lignin is the second most abundant organic substance on earth, and is synthesized in phenylpropanoid pathways. It is known to be composed of three monolignols, that is, p-coumaryl, coniferyl, and sinapyl alcohols. Lignin biosynthesis is thought to occur in three steps: biosynthesis of monolignols, transport of monolignols toward the cell wall, and polymerization of monolignols within the cell wall. Though most enzymes involved in these steps have been identified, their localization in the cell is still obscure. Further investigation of the localization of the enzyme is necessary for better understanding of lignification.
Since PAL was found in barley [1], it has been studied in many herbaceous plants and appears to be pivotal enzyme in the control of the biosynthesis of monolignols [2,3]. PAL is known to be a tetramer in the cell that catalyzes the conversion of l-phenylalanine to cinnamic acid [4]. The molecular mass of the subunit is from 70 to 85 kDa [5–7].
C4H is the enzyme following PAL in cinnamate pathway. C4H catalyzes the conversion of cinnamic acid to p-coumaric acid in the presence of O2 and NADPH [8]. The enzyme exists in the microsomal fraction and is a cytochrome P450-type enzyme [9,10]. The amino acid sequence of C4H indicates that it has the signal peptide on N-terminus targeting to endoplasmic reticulum and combines to the membrane [11]. Moreover, it is reported that the enzymes concerned with phenylpropanoid metabolism form the complex of enzymes and C4H works as the scaffold of the complex [12,13].
In woody stems, PAL- and C4H-GUS activity occurs mainly in differentiating xylem [14,15] and ray parenchyma [16]. Osakabe et al. isolated two PAL genes involved in lignification and demonstrated their expression in the developing xylem but in different stages using immunohistochemistry with the antibody against PAL [17]. Subcellular localization of PAL is observed in the cytosol [18,19], the Golgi apparatus [19], and chloroplasts [20]. Localization of C4H is also observed in the Golgi apparatus [18] and r-ER [13]. Information on subcellular localization of the enzymes involved in lignification is, however, very limited.
We investigated the subcellular localization of PAL and C4H in differentiating xylem of poplar using the antiserums against the synthesized peptides according to the amino acid sequences of PAL and C4H instead of recombinant PAL and C4H proteins. The anti-peptides antibodies are thought to react more specifically to the targeting proteins. The relation between PAL and C4H is also discussed.
2 Material and methods
2.1 Plant materials
Hybrid poplar (Populus sieboldii × P. grandidentata) grown at the nursery of the experimental forests in Kyoto University, Japan was used. For immunostaining, differentiating xylem of poplar was cut into small pieces with a razor blade, soaked immediately in liquid propane cooled with liquid nitrogen, and then immersed into 0.5% glutaraldehyde in acetone cooled at for 3 days for freeze substitution. Thereafter, they were washed with acetone three times, displaced in ethanol, and embedded in LR-White resin (London Resin Co. Ltd., Basingatoke, UK).
For the PAL assay, differentiating xylem of poplar was scraped with a knife, homogenized in liquid nitrogen and extracted in 100 mM Tris-HCl (pH 7.4) containing 10 mM 2-mercaptoethanol, 1 mM EDTA, and 1 mM phenymethylsulfonyl fluoride. The homogenate was centrifuged at for 15 min and the supernatant was centrifuged again at for 20 min using a Microcon YM-10 (Millipore Corporation, Bedford, MA). Then the supernatant was collected and used for PAL assay and Western blot analysis.
For the C4H assay, differentiating xylem of poplar was extracted in phosphate buffer (pH 7.4) containing 250 mM sucrose, 15 mM 2-mercaptoethanol, 1 mM EDTA and 40 mM L-sodium ascorbate. The homogenate was centrifuged at for 15 min, and the supernatant was ultracentrifuged at for 60 min. The pellet was resuspended in 100 mM phosphate buffer (pH 7.4) containing 15 mM 2-mercaptoethanol and ultracentrifuged again at for 80 min. The pellet was resuspended in 100 mM phosphate buffer containing 1.5 mM 2-mercaptoethanol, and used for assay of C4H.
2.2 Preparation of antiserum against PAL and C4H peptides
Whole amino acid sequences of PAL [17] and C4H [21] were used for synthesis of peptides. Three amino acid sequences were selected for epitopes from each amino acid sequence of PAL and C4H using Epitope Adviser software (FQS, Fukuoka, Japan). Because the peptides containing hydrophilic amino acids tend to be exposed on the surface of the native protein, three candidates composed mainly of hydrophilic amino acids were chosen.
Each of these epitopes was composed of 10 amino acids (Table 1). The peptides were synthesized according to the amino acid sequences and conjugated with keyhole limpet hemocyanin (KLH) by Sawady Technology (Tokyo, Japan). The antibodies were raised in mice (6 weeks of age, BALB/C, female). Pre-immune serum was taken from each mouse before the first injection of the antigen. The conjugates were dissolved in phosphate buffered saline (PBS), mixed with the same volume of Freund's adjuvant, and intraperitoneally injected into the mice. The injection was repeated five times at 2-week intervals in the same way, except that Freund's incomplete adjuvant was used. Three days after the last injection, antiserum was taken from the immunized mice, and used for immunolabeling.
Candidates of epitope from whole amino acid sequences of PAL and C4H
| <PAL epitope> | ||
| PAL1 –N terminus–TSHRRTKQGG–C terminus– | (A.A.112∼121) | |
| PAL2 –N terminus–DPLQKPKQDR–C terminus– | (A.A.336∼345) | |
| PAL3 –N terminus–GEKVKSPGEE–C terminus– | (A.A.669∼678) | |
| <C4H epitope> | ||
| C4H1 –N terminus–DRRFESEDDP–C terminus– | (A.A.192∼201) | |
| C4H2 –N terminus–HWKNPEEFRP–C terminus– | (A.A.409∼418) | |
| C4H3 –N terminus–QSJIDTSEKG–C terminus– | (A.A.476∼485) |
2.3 Dot blot and Western blot assay
The specificity of antiserums against PAL and C4H peptides was examined by dot blot and Western blot assay. For dot blot, 2 μl of KLH solution, peptides conjugated with KLH, and crude enzyme extracts from differentiating xylem of poplar were spotted on sheets of nylon membrane (Biodine B/Pall, East Hills, NY). The sheets were incubated with blocking buffer containing 3% skim milk in PBS. The antiserums against PAL and C4H peptides, the antiserums pre-incubated with KLH, and pre-immune serum were diluted 1:200 with 3% skim milk in PBS, and were used to incubate the sheets. Then sheets were incubated with alkaline phosphatase-conjugated goat antiserum against mouse IgG (ICN Pharmaceuticals, Aurora, OH). The antiserum was detected by p-nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP). For Western blot, the crude enzymes were charged to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) performed with 10% polyacrylamide gels. The separated proteins were transferred to PVDF membrane (Bio-Rad, Hercules, CA). After incubation with antiserum against PAL and C4H peptides, both membranes were stained according to the same procedure for dot-blot assay.
2.4 Immuno-inhibition test of PAL
PAL activity was measured by using the crude enzymes extracted from the differentiating xylem of poplar. For immuno-inhibition assay, the enzymes solution was pre-incubated with the antiserum against PAL peptides or pre-immune serum for 30 min, followed by incubation with Magna Bind anti-mouse IgG (PIERCE, Rockford, IL) for 15 min at room temperature. The protein concentration of antiserums was checked by the Bradford assay monitored at 595 nm using a spectrophotometer (UV-1600; SHIMADZU, Japan). After centrifugation, the supernatants were mixed with 20 mM L-phenylalanine in 100 mM phosphate buffer (pH 7.4), and incubated for 60 min at 35 °C. The reaction was stopped by adding 5 N HCl. Thereafter trans-cinnamic acid in the reaction mixtures was monitored at 268 nm. The reaction mixture without l-phenylalanine was also measured as a control.
2.5 Immuno-inhibition test of C4H
C4H activity was measured using a microsomal fraction of poplar. Microsomal solution (50 μg ml−1) was incubated with the antiserum against C4H peptides followed by incubation with Magna Bind anti-mouse IgG. C4H activity was measured according to the method of Lamb and Rubery [22]. After the supernatant was mixed with 200 μM trans-cinnamic acid, 500 μM NADP, 1U of glucose-6-phosphate dehydrogenase, 10 mM d-glucose-6-phosphate monosodium salt, and 100 mM phosphate buffer (pH 7.4), the mixture was incubated for 30 min at 35 °C. The reaction was stopped by adding 5 N HCl. The solution was centrifuged at for 10 min and the supernatant was adjusted to pH 11 with NaOH. Thereafter, p-coumaric acid in the reaction mixture was determined at 340 nm using a spectrophotometer. The reaction mixture without trans-cinnamic acid was also treated in the same manner as a control.
2.6 Immunofluorescence microscopy
Thin sections (1 μm thick) were cut from the embedded block and mounted on glass slides. The following immunolabeling procedures were performed at room temperature. The sections were incubated for 15 min with 50 mM glycine in PBS (pH 7.4) followed by incubation for 30 min with 3% skim milk in PBS to avoid nonspecific binding of antibody. They were then incubated with the antiserum against the peptides of PAL or C4H (diluted 1:100 in 3% skim milk in PBS) for 2 h. After washing with PBS, they were incubated with fluorescein isothiocyanate-labeled goat anti-mouse IgG antibody (Zymed laboratories, Inc., San Francisco, CA) for 1 h. They were again washed with PBS, treated with 2% glutaraldehyde in PBS for 5 min, washed with distilled water, and observed under a confocal laser scanning microscope (FLUOVIEW FV300; Olympus, Japan).
2.7 Immunogold labeling for electron microscopy
Ultra-thin sections mounted on formvar-coated nickel grids were used. The following immunolabeling procedures were performed at room temperature unless otherwise specified. The sections on grids were soaked in 50 mM glycine in PBS (pH 7.4) for 15 min, and then incubated in blocking buffer (PBS containing 0.8% BSA, 0.1% IGSS-quality gelatin [Amersham, Little Chalfont, UK], 5% goat serum, and 2 mM NaN3) for 1 h. After washing three times with washing buffer (same as blocking buffer, but without goat serum) for 10 min, they were incubated with the antiserum against peptides of PAL or C4H (diluted 1:100 in blocking buffer) overnight at 4 °C. After washing three times with a washing buffer for 10 min, they were incubated with 15-nm colloidal gold-conjugated goat anti-mouse IgG antibody (Auro Probe EM, GAR15; Amersham; diluted 1:25 in blocking buffer) for 90 min. They were washed three times for 10 min with washing buffer, then washed with PBS, treated with 2% glutaraldehyde in PBS for 5 min and washed with distilled water. Control sections were treated in the same way, with the exception that mouse pre-immune serum was used. The sections were stained with 2% aqueous uranyl acetate followed by staining with Reynolds' lead citrate, and then examined under a transmission electron microscope (JEM 1220; JEOL, Tokyo, Japan) at 100 kV.
3 Results
3.1 Dot blot assay
The results of the dot blot assay are shown in Fig. 1. Each antiserum against PAL peptides reacted with KLH, each PAL peptide-KLH conjugate, and crude enzymes extracted from differentiating xylem of poplar. The antiserums against PAL peptides pre-incubated with KLH did not positively react with KLH, but reacted with PAL peptide-KLH conjugates and the crude enzymes. The antiserums against C4H peptides pre-incubated with KLH also positively reacted with C4H peptide-KLH conjugates and the crude enzymes. Pre-immune serums did not react with KLH, PAL and C4H peptide-KLH conjugates, or the crude enzymes.
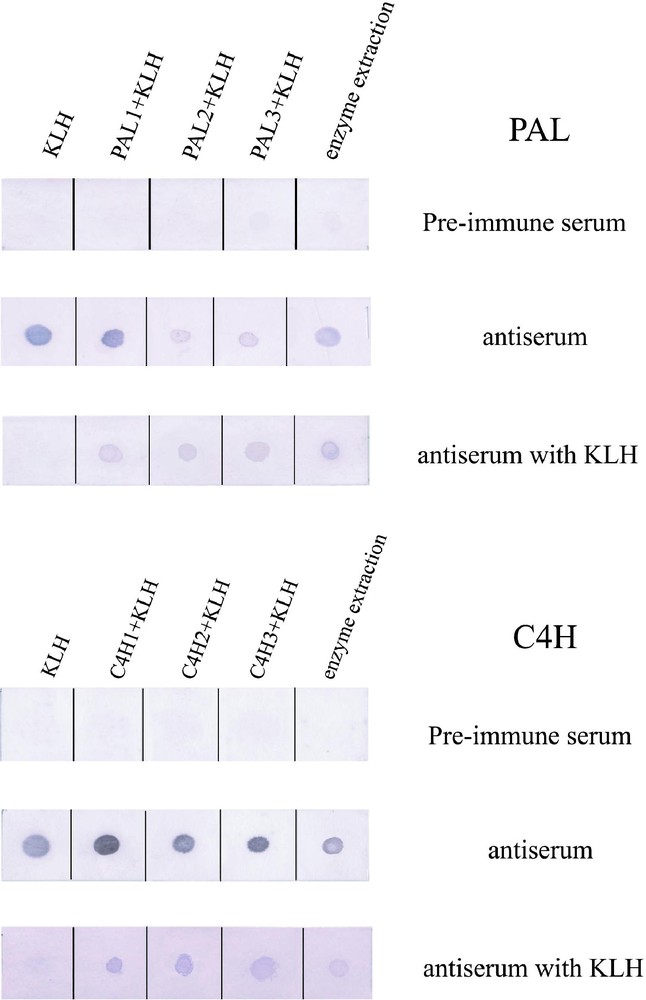
Dot blot assay by using antiserums against PAL and C4H peptides. KLH, peptides conjugated with KLH and the crude enzymes extracted from the differentiating xylem of poplar were spotted from the left side of the membrane.
3.2 Western blot assay
The antiserum against PAL2 peptide reacted only with a 75-kDa protein among the crude extracts from differentiating xylem of poplar. However, the antiserums against PAL1 and PAL3 peptides did not show any reaction to the crude extracts. The antiserum against C4H3 reacted only with a 58 kDa protein (Fig. 2), but those against C4H1 and C4H2 did not show any reaction. The putative molecular weights of PAL and C4H are 77 kDa and 58 kDa, respectively, according to the total molecular mass of amino acids. Pre-immune serum did not react with crude enzymes (data not shown). Therefore, the antiserums raised against PAL2 and C4H3 peptides were used in the following experiments.
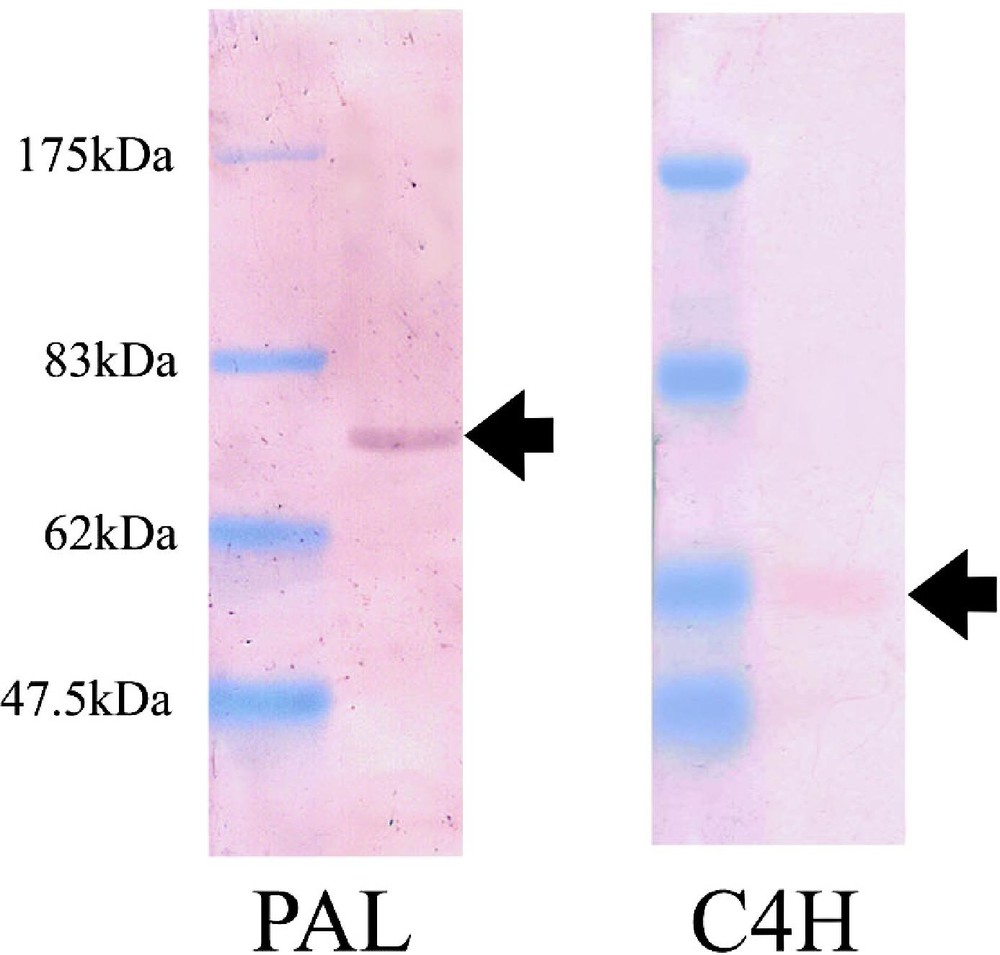
Western blot assay by using the antiserums against PAL2 and C4H3 peptides. Both antiserums reacted to a single band from the crude enzyme extraction. The antiserums of PAL- or C4H-peptide recognized specifically 75 kDa and 58 kDa polypeptides, respectively.
3.3 Immuno-inhibition test of PAL and C4H
When the antiserum against PAL2 peptide was added in the crude enzyme extract from poplar with phenylalanine, absorbance at 268 nm decreased with an increase in antiserum. The addition of the antiserum against C4H3 peptide to the microsomal enzyme extract with trans-cinnamic acid also caused a decrease in the 340-nm absorbance (Fig. 3). These results indicated that antiserums of PAL and C4H peptides inhibited the conversion of phenylalanine into trans-cinnamic acid, and that of trans-cinnamic acid into p-coumaric acid, respectively. These antiserums might specifically bind to PAL and C4H protein, respectively.
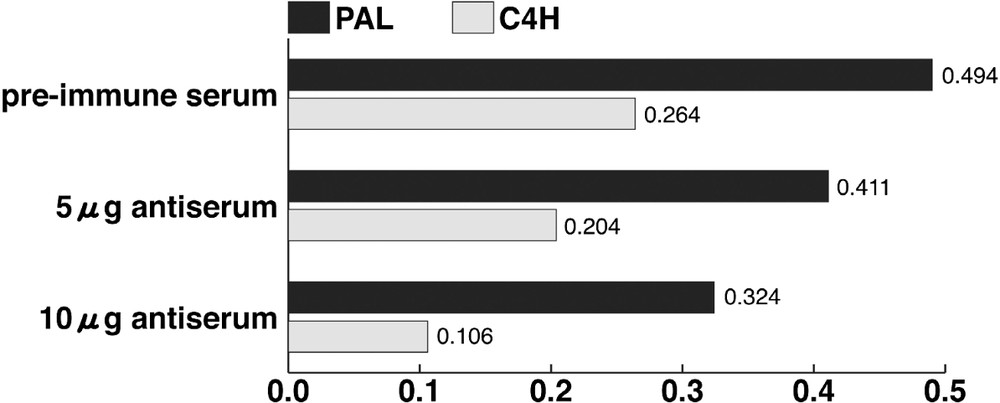
Immuno-inhibition test of PAL and C4H. Horizontal axis shows the absorbance at 268 and 340 nm for measuring PAL and C4H activities, respectively.
3.4 Immunolocalization of PAL and C4H
Immunofluorescence microscopy revealed that PAL immunolabeling was localized in the differentiating xylem of poplar. In particular, the labeling was strong from the late stage of primary cell wall formation to the early stage of secondary cell wall formation (Fig. 4A). Intense labeling was also observed in some of ray parenchyma cells. Immunolabeling of C4H was observed in the early stage of secondary cell wall formation. The labeling was, however, observed as spots in the cells (Fig. 4B). Under a transmission electron microscope, immunolabeling of PAL was mainly distributed in the cytosol in differentiating xylem cells (Fig. 5A and B). Weak labeling was observed on the developing secondary cell walls, r-ER, plastids, and vacuoles. In contrast to the PAL immunolabeling, the C4H labeling was observed on r-ER and the Golgi apparatus in the differentiating fiber and vessel (Fig. 5C and D). It was not observed, however, in cytosol as much as PAL immunolabeling. The labeling was not observed when the section was treated with the pre-immune serum (Fig. 5E and F).
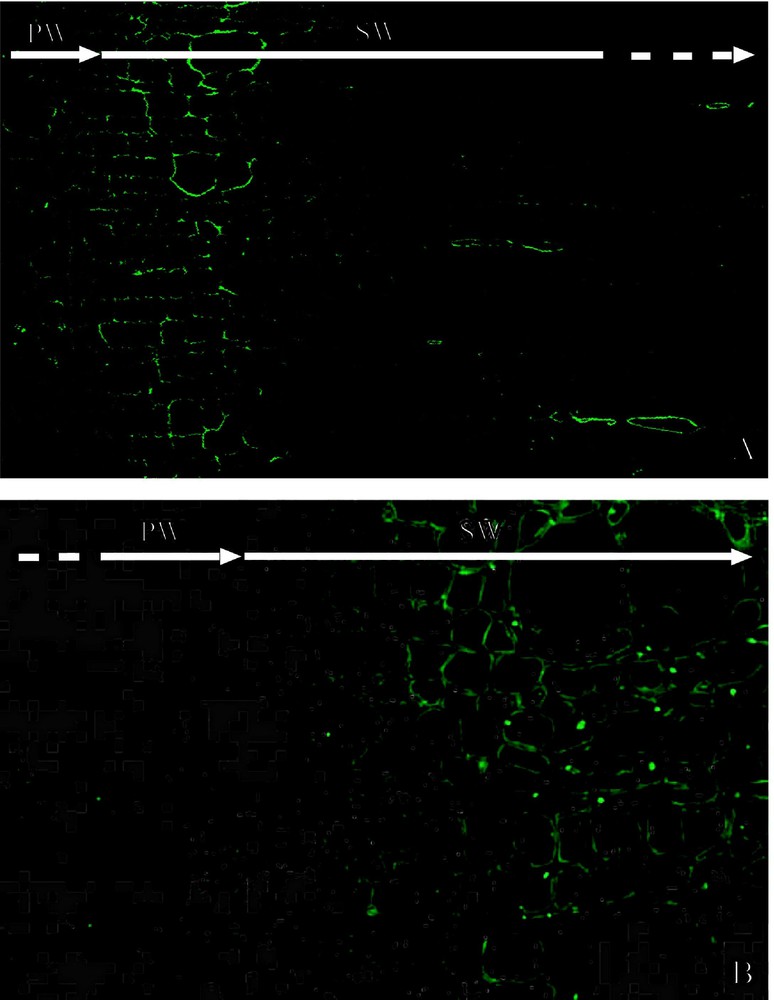
Immunofluorescence microscopic observation of PAL and C4H in the differentiating xylem of poplar. (A) Immunolocalization of PAL. (B) Immunolocalization of C4H. PW: Primary cell wall formation. SW: Secondary cell wall formation.
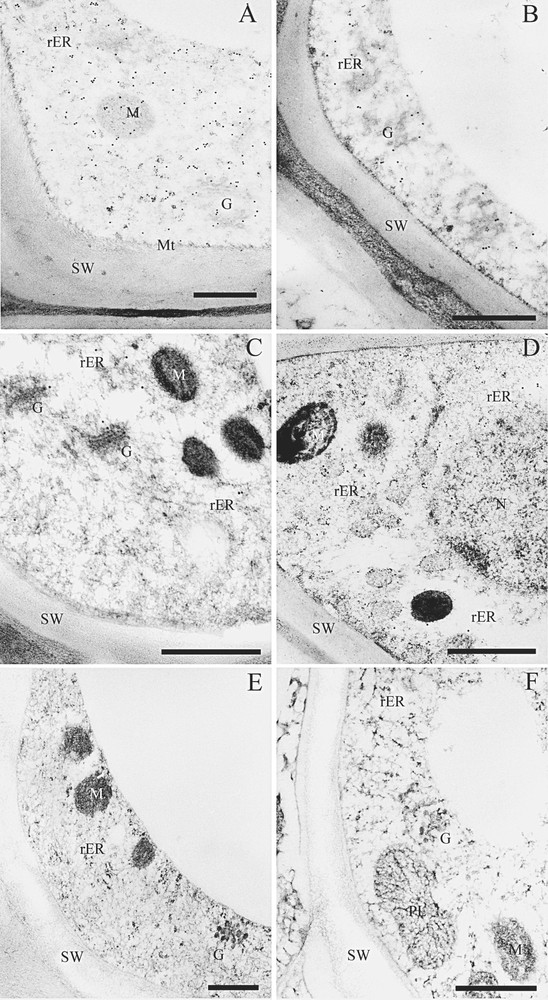
Immunogold labeling of PAL and C4H in the differentiating xylem. (A) Immunolocalization of PAL in the differentiating fiber. (B) Immunolocalization of PAL in the differentiating vessel. (C) Immunolocalization of C4H in differentiating fiber. (D) Immunolocalization of C4H in the differentiating xylem. (E) Control photo by using the pre-immune serum. (F) Control photo by using the pre-immune serum. Bar = 500 nm; G, the Golgi apparatus; M, mitochondria; Mt, microtubules; N, nucleus; PL, plastid; rER, rough-endoplasmic reticulum; SW, secondary cell wall.
PAL and C4H immunolabeling was determined by counting the gold particles on the 20 electron photomicrographs (Fig. 6). Total numbers of PAL and C4H labeling were 836 and 561, respectively. More than half of the PAL labeling was localized in the cytosol, though the labeling on r-ER and the Golgi apparatus were 8 and 13%, respectively. In contrast to PAL, only 20% of the C4H labeling was localized in the cytosol. Total amount of the labeling on r-ER, the Golgi apparatus, and plastids was approximately 80%.
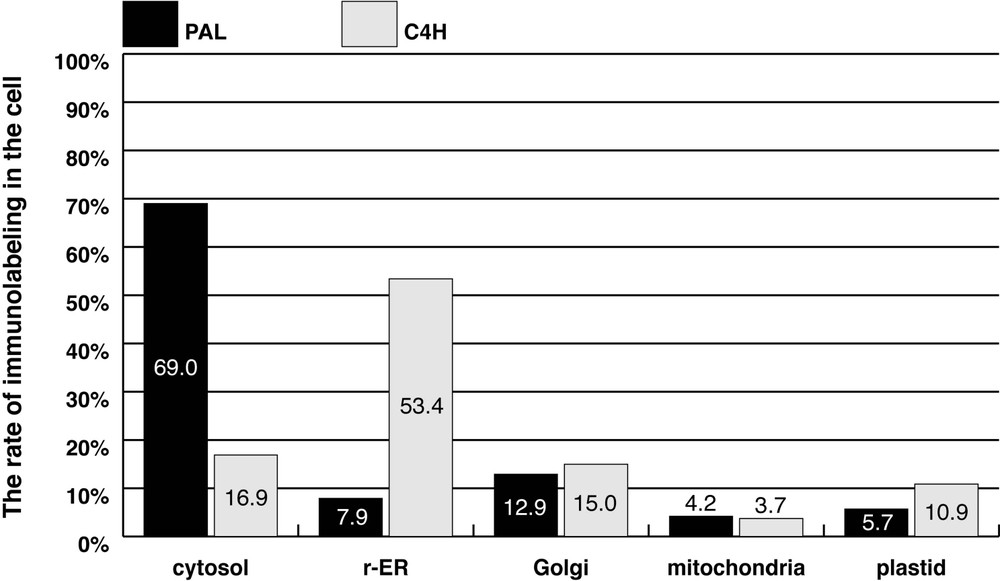
Semi-quantitative analysis of PAL and C4H immunolabeling.
4 Discussion
4.1 Characterization of antiserum
The dot blot immunoassay clearly indicated that the antiserums against the PAL and C4H peptides contained antibodies against the corresponding peptides. Because these antiserums react positively to the crude enzymes extracted from the differentiating xylem of poplar, the enzymes contain proteins with the same amino acid sequences of PAL and C4H. Western blot assay indicated that the antiserum against the PAL peptide recognized only the 75-kDa protein in the crude enzyme extract. The molecular mass of the recognized protein corresponds to the molecular mass of the PAL subunit [17]. The antiserum against the C4H peptide recognized protein with a molecular mass of 58 kDa, which corresponds to that of C4H [10]. The result of the inhibition assay of PAL and C4H indicated that both activities decreased gradually with an increase in PAL and C4H antiserums, respectively. The above results indicate that the antiserums against the PAL and C4H peptides contain antibodies that bind specifically to PAL and C4H, respectively.
4.2 Localization of PAL and C4H in the cell
PAL immunolabeling in differentiating xylem of poplar suggests that the conversion of phenylalanine to cinnamic acid occurs in this region. Osakabe et al. showed that PAL was specifically localized in differentiating xylem by immunohistochemistry using the antiserum derived from the PAL recombinant protein [17]. Subramaniam et al. reported that PAL gene expression was observed in young stem and lower part of old stem [23]. Our result supports the above results. Intense PAL immunolabeling in ray parenchyma cells might be involved in not only the synthesis of monolignols but also in the synthesis of other phenolic substances. Further investigation is needed to reveal the function of ray parenchyma cells. Transmission electron microscopic observation indicated that most of the PAL immunolabeling is localized in the cytosol during secondary wall formation. Weak labeling was also observed on r-ER and the Golgi apparatus.
C4H immunolabeling is localized in the differentiating xylem in poplar. However, the labeling is observed as a spot, though the labeling of PAL is homogeneously distributed in the cytosol. Electron microscopic observation indicated that most C4H labeling is localized on r-ER and the Golgi apparatus. The labeling is also observed on plastids and mitochondria. Semi-quantitative analysis of C4H labeling demonstrates that the majority of the labeling is localized on r-ER, and some on the Golgi apparatus. Hydrophobic plot analysis strongly suggests that C4H has a signal peptide at the N-terminus and a single transmembrane domain near the N-terminus (not shown). Therefore, C4H might be localized on the membrane of r-ER and the Golgi apparatus. Ro et al. reported that C4H is localized in r-ER [13]. Our results support their results. Chapple suggested that the catalytic domain of C4H was localized at the cytosolic site [24]. The conversion of cinnamic acid to p-coumaric acid might occur at cytosolic site of the membrane of r-ER and the Golgi apparatus.
Rasmussen and Dixon showed that total activity of PAL in microsomal fraction from wild-type tobacco plants amounted to 5 to 10% of the total activity of PAL in the soluble fraction. In addition, the conversion of phenylalanine to p-coumaric acid via cinnamic acid occurred rapidly in their in vitro experiment [12]. Therefore, they proposed that one or more PAL isoforms are associated with C4H as an enzyme complex on r-ER. Our results indicate that PAL and C4H are localized simultaneously in the differentiating xylem in poplar. Some PAL labeling is localized on r-ER and the Golgi apparatus, and there is much C4H labeling on r-ER. These results suggest the possibility of coordinated localization of both enzymes. Double PAL and C4H immunolabeling in the same section will provide more information on their relation in the cells.
Acknowledgments
The authors are very grateful to Dr H. Ebinuma and Dr K. Watanabe at Nippon Paper Industries in Japan, and to Dr S. Inoue at Akita Jujo-Kasei Industries in Japan for providing us the poplar.


