1 Introduction
Translational control is critical for the proper regulation of cell cycle, tissue induction and growth, normal embryogenesis, and germ-line development [1–5]. Translational regulation of an eukaryotic mRNA is achieved through the orchestrated action of cis-acting elements and trans-acting factors. Cap-dependent translation in eukaryotes requires the ordered assembly of a complex of evolutionarily conserved proteins, which starts with the binding of the translation initiation factor 4E (eIF4E) to the 7-methyl-guanosine (m7GpppN) cap structure at the 5′ of the mRNA. Next, the eIF4G factor is recruited allowing additional factors (PABP, eIF4A, eIF4B, eIF1, eIF1A, eIF2, eIF3, among others, and the ribosomal subunits) [6] to form a complex that, after mRNA circularization, initiates translation [7–10]. The circularization might become possible once an adequate poly(A) tail is present at the 3′-UTR [11].
eIF4E is the rate-limiting component for cap-dependent translation initiation and therefore represents a major target for translational control [12,13]. eIF4E function can be regulated at different levels by a variety of molecular processes. First, eIF4E transcription inside a cell can be increased by growth factor stimuli [14,15]; second, the availability of eIF4E can be modulated by the binding to a set of proteins that compete with eIF4G, a scaffold protein that aggregates the mRNA and the ribosome [16], for eIF4E-binding, therefore inhibiting translation initiation [17]; third, elevated eIF4E activity depends upon its phosphorylation in response to extracellular stimuli, including hormones, growth factors, and mitogens, and correlates to an increase in translation rate [16,18]. The function of phosphorylated eIF4E was indeed shown to be necessary for proper growth and development of Drosophila [19].
During oogenesis in many species, cytoplasmic polyadenylation of a set of maternal mRNAs regulates their translation. A general scheme implies that elongation of a short poly(A) tail at the 3′-UTR during development is able to stimulate translation. The cytoplasmic polyadenylation element binding protein (CPEB) is a sequence-specific RNA interacting factor that is necessary to achieve adequate poly(A) addition within the cytoplasm. The rationale for the existence of a long poly(A) tail is, probably, to allow an mRNA to acquire a circular structure prior to translation initiation. Increasing evidence indeed suggest that the structure of an mRNA is essential for proper activity, including translation efficiency.
The past few years have witnessed considerable advancements in the field of translational control during development. In particular, recent studies have revealed the existence of various translational regulatory mechanisms and identified in the cap-poly(A) tail interaction the primary target for multiple regulatory factors. Moreover, novel repressive and stimulatory complexes involved in translational regulation of specific mRNAs have been described and analyzed.
In this review, we will focus on the molecules that by binding and/or modulating the modifications occurring at the end of mRNAs, the cap structure at the 5′ and the poly(A) tail at the 3′ end, are the targets of regulatory events that govern translation of select mRNAs playing crucial roles in specific developmental processes.
2 Structure of eukaryotic mRNAs and translational control
In the nucleus, eukaryotic mRNAs are first transcribed as precursor mRNAs (pre-mRNAs) and subsequently modified by capping, polyadenylation, and splicing. Mature mRNAs are ultimately exported into the cytoplasm where they can be translated into proteins.
Capping of eukaryotic mRNAs involves the addition of a 7-methyl-guanosine residue at the 5′ end to protect this end from nuclease degradation. The cap structure in eukaryotes can be of three types m7GpppNp, m7GpppNmp, m7GpppNmpNmp (m indicates a methyl group attached to the respective nucleotide), and is used as a docking point for the cap-binding protein complex that mediates the recruitment of the small ribosomal subunit to the 5′ end of the mRNA.
Polyadenylation occurs after cleavage of the pre-mRNA at the 3′ end and consists of the addition of up to 250 adenosine residues by the poly(A) polymerase (PAP) enzyme. Finally, the mechanism of splicing removes all intervening sequences from the pre-mRNA, thus producing a mature mRNA that is competent to be transported to the cytoplasm.
Once within the cytoplasm, only mRNAs that are properly capped and polyadenylated are efficiently translated. This has been demonstrated during oogenesis and early embryogenesis, where regulated cytoplasmic polyadenylation of mRNAs modulate their translation. During translation initiation, the cap structure is directly bound by eIF4E, and the poly(A) tail by the poly(A) binding protein (PABP) in a manner that induces a synergistic enhancement of translation. Translation that is both cap- and poly(A)-dependent requires moreover the activity of the eukaryotic initiation translation factor 4G (eIF4G). eIF4G contains specific binding sites for both eIF4E and PABP, thus forming a complex that circularizes the mRNA [10]. Indeed, mutations that compromise the binding of eIF4G with either PABP [20,21] or eIF4E [22,23] affect in vitro the synergistic stimulation of translation by the cap structure and the poly(A) tail. In this context, the 5′ cap and the 3′ poly(A) tail are modifications necessary for efficient translation.
Regulatory sequences and structures within the mRNA modulate translation. In addition to the 5′ cap and 3′ poly(A) tail these include: internal ribosome entry sites (IRESs), which direct cap-independent translation initiation; upstream open reading frames (uORFs), which act as negative regulators by diminishing translation from the main ORF; secondary or tertiary structures, like hairpins and pseudoknots, which often act by blocking translation initiation; and specific binding sequences for multiple regulatory factors [6]. Many mRNAs are translationally controlled by sequences in their 5′ and 3′ untranslated regions (UTRs). Embedded within untranslated regions of eukaryotic mRNAs are information specifying the way the RNA is to be utilized and diverse proteins bind specifically to these sequences thus interpreting this information [24].
3 Eukaryotic translation initiation factor 4E (eIF4E) and cap-binding complex assembly
The recruitment of the small ribosomal subunit to mRNA is one of the tightly regulated steps in the initiation of protein synthesis. Two major pathways are involved in the attachment of the small ribosomal units 5′ to the translated region in mRNAs: the first depends on the 5′ terminal cap structure of mRNAs, consisting of an inverted methylated guanine moiety (m7GpppN); the second, known as cap-independent, relies on a series of elements of complex secondary structure, present in select mRNAs, termed IRES (internal ribosomal entry sites). The vast majority of eukaryotic transcripts are translated in a cap-dependent manner.
The cap structure represents a docking point for the cap-binding protein complex and is required for binding of the small ribosomal subunit to the 5′ end of mRNAs. This protein complex has three main components, the cap-binding protein eIF4E, the adaptor protein eIF4G, and the poly(A)-binding protein (PABP). The direct interaction of eIF4E, bound to eIF4G, to the cap structure is essential for translation initiation both in vivo and in vitro, while contacts between eIF4G and the poly(A)-bound PABP enhance translation (Fig. 1A), but are not strictly required for ribosome recruitment (see reviews [25–27]). Thus, events favouring the dissociation of the eIF4E–eIF4G interaction significantly impair cap-dependent translation and are therefore a potential means for translational control. Such a control has indeed been observed both in developmental processes and in tumorigenesis [28,29].
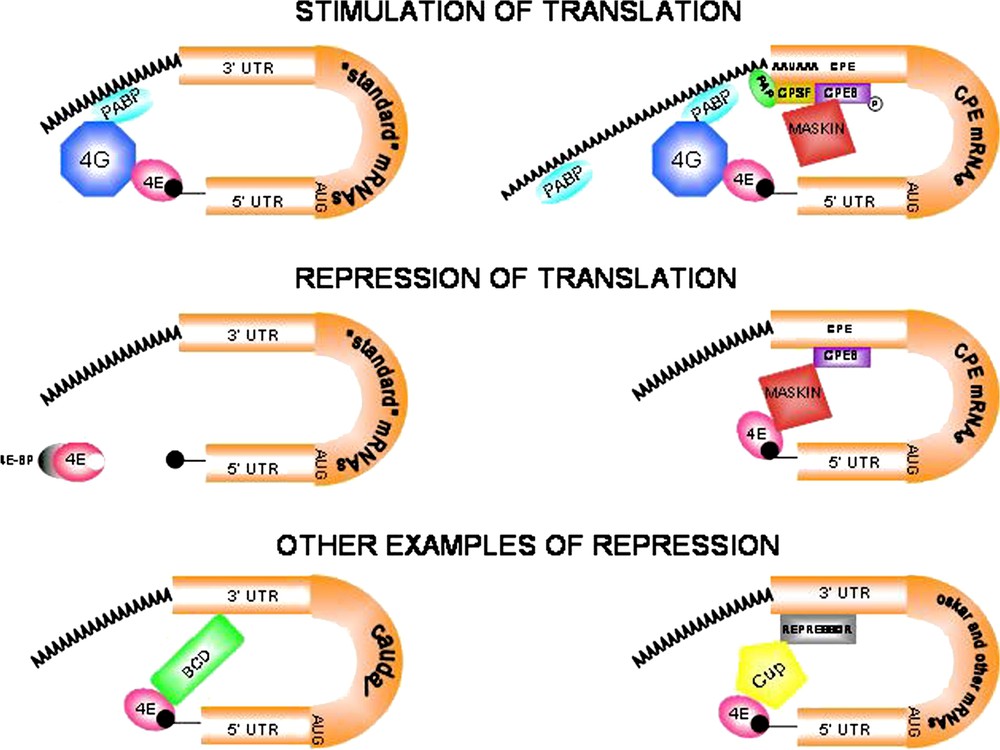
Modes of translational regulation during oogenesis and early development. (A)–(B). Stimulation of eukaryotic mRNA translation. (A) ‘Standard’ mRNAs. The poly(A) binding protein (PABP) binds the scaffolding protein eIF4G (4G) that in turn interacts with eIF4E (4E), thus promoting mRNA circularization and translation. (B) CPE-containing mRNAs are activated by cytoplasmic polyadenylation. The cytoplasmic polyadenylation element binding protein (CPEB) binds specific U-rich sequences named cytoplasmic polyadenylation elements (CPEs). Phosphorylated CPEB increases its affinity for the cleavage and polyadenylation specificity factor (CPSF). CPSF in turn interacts with the canonical nuclear polyadenylation signal AAUAAA and recruits poly(A) polymerase (PAP) to the 3′ end of the mRNA. The consequent poly(A) tail elongation stimulates translation of dormant mRNAs. The AAUAAA sequence is written 5′ to 3′ for clarity. (C)–(F) Repression of eukaryotic mRNA translation. (C) ‘Standard’ mRNAs. eIF4E-binding proteins (4E-BPs) are inhibitory proteins that regulate the availability of eIF4E for interaction with eIF4G. 4E-BPs sequester eIF4E molecules that are either free or cap-bound. Hyperphosphorylation of 4E-BPs prevents interaction with eIF4E, thus allowing interaction with eIF4G and translation to start (see panel A). (D) CPEB mediates both activation and repression of translation. The repressive role of CPEB involves the interaction with Maskin. Maskin associates with the translation initiation factor eIF4E and excludes eIF4G from interacting with eIF4E, thus blocking initiation of translation of CPE-containing mRNAs. The Maskin-eIF4E complex is disrupted by cytoplasmic polyadenylation triggered by CPEB, allowing eIF4G to bind eIF4E and activate translation. Translation repression is relieved when PABP binds to the poly(A) tail and helps eIF4G displace Maskin and bind eIF4E (see panel B). (E) Bicoid (BCD) is a specific repressor of caudal (cad) mRNA translation. The anterior determinant BCD protein acts not only as a transcriptional activator of segmentation genes, but also interacts directly with both the 3′-UTR of caudal mRNA and eIF4E to disrupt the eIF4E-eIF4G complex, thus preventing translation initiation of ubiquitously distributed cad mRNA. (F) Translational inhibition of oskar (osk) and other mRNAs. Cup is a translational regulator that acts, by interacting simultaneously with eIF4E and RNA-bound repressor molecules, in translational inhibition of specific mRNAs. Accordingly, two repressors, Bruno and Smaug, suppress translation of osk and nanos (nos) mRNAs respectively by binding to their 3′-UTRs prior to their posterior localization in the egg (osk) and embryo (nos).
The functions of eIF4E and the regulation of these depend on the presence on this factor of binding surfaces for the cap-structure of mRNAs and for various proteins that can modulate its activity. The three-dimensional structure of the eIF4E-cap complex has been solved, identifying the relevant molecular contacts between the 5′ end of mRNAs and eIF4E [30–35]. The cap-binding site of eIF4E was shown to be formed by a pocket, which contains two critical tryptophan amino acid residues located close to its upper and lower edges. The guanosine moiety was shown to make contacts with these tryptophan residues. Moreover, the presence of a methyl group, by introducing a positive charge, was predicted to enhance considerably the interaction [34,36–42]. No relevant contacts beyond the ones with the first nucleotide have been detected, indicating that the molecular interactions of eIF4E with the cap are essentially identical regardless of the mRNA species. Thus, since the methylated guanosine is present on all capped mRNAs, it can be excluded that transcript-dependent efficiency of translation is obtained by specific contacts between eIF4E and nucleotides other than the cap.
Some evidence was presented for the existence of conformational changes of eIF4E upon cap binding [43–45], suggesting that some or all of the factors interacting with eIF4E might be able to discriminate between cap-bound and apo eIF4E. One important example for changes in the strength of protein–protein contacts is represented by the interaction with the inhibitors 4E-binding proteins (4E-BPs), which was shown to be significantly increased for the cap-bound form of eIF4E [46]. A further example is provided by the interaction with eIF4G. eIF4E forms a very stable complex with eIF4G. The eIF4G-binding surface within eIF4E is located distally to the cap-binding pocket and does not contain any known residues involved in cap binding [32,47]. NMR studies have shown that upon interaction eIF4E and eIF4G mutually induce conformational changes that result in a complex interlocking structure [30]. Binding of eIF4G to human eIF4E is mediated by the dorsal region of eIF4E containing a Trp residue, which is also required for interaction with 4E-BPs [32,39]. 4E-BPs are small proteins showing no defined structure in solution, which undergo conformational changes in the regions contacting eIF4E. All proteins known to interact with eIF4E bind to a common region located distally to the cap-binding pocket even if their contacts with eIF4E, which are not yet fully characterized, may vary considerably. The interaction of eIF4E with the cap structure is considerably stabilized after eIF4G binding. The RNA-binding activity of eIF4G has been invoked as an explanation for this. However, possible conformational changes occurring upon eIF4E–eIF4G interaction that stabilize cap-binding have also to be considered. A combination of both mechanisms could indeed account for the enhanced stability of the eIF4E–eIF4G–PABP complex to the mRNA 5′ end. The notable conformational changes, which occur upon eIF4G-binding to eIF4E, were shown to induce changes in the structure of the cap-binding site of eIF4E [30]. Binding of the PABP protein was shown to further stabilize the cap interaction [45,48–51]. PABP binding to eIF4G is enhanced by contacts with the poly(A) tail and RNA maximal cap-binding activity likely depends upon the formation of a full eIF4E–eIF4G–PABP–poly(A) complex.
3.1 Regulation of translation via competition of the eIF4E–eIF4G interaction
In addition to the 4E-BPs several other eIF4E-binding proteins have been identified in a number of different species, which can compete for the assembly of a translationally active eIF4E–eIF4G complex and hence act as regulators of translation (Table 1). These are usually specialized proteins that play specific roles during developmental processes. The best characterized examples are the X. laevis Maskin [52], the Drosophila Bicoid (BCD) and Cup proteins [53–56]. These proteins share with eIF4G and the 4E-BPs the characteristic motif Tyr–X–X–X–X–Leu–Φ (where X represents any residue and Φ is leucine, methionine or phenylalanine), which is required for interaction with eIF4E and was hence termed eIF4E-binding motif. Three basic modes of action have been described for proteins sharing an eIF4E-binding motif that act antagonistically on the formation of an eIF4E–eIF4G complex. The first is represented by the 4E-BPs, which act as general repressors of translation in response to cell growth-dependent signalling events (Fig. 1C) [8,9]. The second is exemplified by the Maskin protein, which acts in an mRNA-specific manner. The target specificity of repression is obtained by the interaction of Maskin with the cytoplasmic polyadenylation binding protein (CPEB) that in turn recognizes a uridine-rich sequence (CPE) present in the 3′-untranslated region (3′-UTR) of selected mRNAs [24,57]. The repression itself is brought about by a direct Maskin-eIF4E interaction (Fig. 1D) [58,59]. The third mechanism is represented by the BCD protein which can both directly interact with a specific mRNA (caudal mRNA) and disrupt the eIF4E–eIF4G complex (Fig. 1E) [60]. Recently an additional eIF4E-interacting protein, Cup, was identified in Drosophila and shown to play crucial roles in translational control and in the localization of eIF4E during Drosophila oogenesis and ovary development [53,55,56]. While the capability of Cup to specifically repress translation has not been experimentally verified, the findings that Cup is able to antagonize binding of eIF4G to eIF4E [54,56], and that Cup is associated to specific mRNAs, which are transported during oogenesis and whose translation is blocked during transport [53,55], would suggest that Cup negatively controls translation as well (Fig. 1F). Cup has been suggested to act via a mechanism similar to that of Maskin [61]. There is evidence that Cup is recruited to the oskar (osk) and nanos (nos) mRNAs by the Bruno and Smaug proteins, respectively, which specifically contact these mRNAs (Fig. 1F) [53–55]. However, Cup might play a role in osk translational repression also independently of its binding to osk mRNA since the loss of Cup activity was shown to have a more pronounced effect on osk translation than preventing Bruno from binding to osk mRNA. Moreover, Cup was shown to be associated with Barentz, a protein required for the localization of the osk mRNA, indicating that Cup plays a role in the localization of the osk mRNA within the oocyte as well [55]. Interestingly, the localization and the overall amount of the eIF4E protein, which accumulates at the posterior end of the developing Drosophila oocyte, are altered in a cup mutant background [55,56], further suggesting that Cup plays more general functions in translational regulation within the oocyte, likely by controlling the translation of a number of different mRNAs. Indeed, mutation of cup was shown to have a substantial effect on oocyte maturation and ovary development [62,63], and a reduction of eIF4E levels within ovaries causes a significant aggravation of the cup mutant phenotype [56]. Thus, the role of Cup might be to coordinate localization of a number of mRNAs during Drosophila oogenesis and to assure their translational repression during transport. These transcripts might encode products asymmetrically distributed within the oocyte to assure its proper maturation.
Putative functions of eIF4E interactors in translation
| Interactors | Function of the interactions in translation |
| eIF4G's | Stimulation of translation of all mRNAs |
| Maskin | Repression of translation of CPE-containing mRNAs |
| Bicoid | Repression of translation of caudal mRNA |
| Cup | Repression of translation of oskar and other mRNAs |
| 4E-BP's | Repression of translation of all mRNAs |
| Emx2 | Local control of translation in olfactory sensory neurons |
| HOXA9 | Stimulation of the nuclear export of cyclinD1 and ODC mRNAs |
3.2 eIF4E levels and translation
A crucial aspect of the regulation of translation initiation is the relationship between eIF4E levels and translational activity within cells. Changes in the abundance or activity of eIF4E have been reported in several cases and linked to tumorigenesis [29], adaptation to environmental stresses [64], and developmental processes [28]. The relative amount of eIF4E and other translation initiation factors has been a controversial issue. It has been initially reported that eIF4E levels are limiting, at least in mammalian cells and reticulocyte lysates [65,66]. However, more recent data indicate that eIF4E is present in reticulocyte lysates in excess over eIF4G, and that in yeast it is equimolar to ribosomes and to other initiation factors. Thus, apparently, in reticulocytes, yeast, and D. melanogaster cells, it is the availability of eIF4G, rather than that of eIF4E, which limits the frequency of translation initiation [34,45,67,68]. The availability of eIF4E could be modulated by the activity and/or abundance of 4E-BPs within cells. Under active growth conditions the 4E-BPs are in a hyperphosphorylated state, due to the activity of the FRAP/mTOR kinase [69], thus inhibiting binding to eIF4E [70,71]. In these conditions, eIF4E is likely present predominantly as a complex with eIF4G or in the apo eIF4E form. When cells are not actively growing and hence the levels of protein synthesis are reduced, the 4E-BPs remain in a dephosphorylated state and thus are able to bind eIF4E, thus reducing the availability of free eIF4E. The extent of 4E-BPs phosphorylation and the efficiency of competition between eIF4E–4E-BP and eIF4E–eIF4G complexes for cap-binding would determine the levels of translational activity within cells.
Experimental variations of eIF4E levels in vivo have proven to be valuable in estimating the stability of the translation initiation complex within cells. The overexpression of eIF4E by up to a factor 100 in yeast has only minor effects on growth rates [72], while a modest increase in translation rates could be observed in X. laevis cells [73]. Thus the overall translation levels are apparently not significantly affected by increases in eIF4E availability. Along these lines, a 30% reduction of eIF4E wild-type levels is tolerated in yeast without apparent effects on growth rate; and translation in yeast is seemingly affected only as soon as eIF4E levels fall close to or below those of total mRNA [45]. Thus, at least under conditions of high translational activity, eIF4E levels do not seem to influence bulk translation. Nevertheless, alterations of eIF4E levels that are supposed to leave bulk translation unchanged can have profound effects on the physiology of cells. For example, in yeast, reductions in eIF4E levels that do not impair general translation, significantly affect cell morphology, ribosome biogenesis and cell cycle progression [45,74]. The importance of eIF4E levels for the translation of select mRNAs has been recently shown in the case of the Drosophila neuromuscular junctions (NMJ). eIF4E accumulates at Drosophila larval NMJs to sustain local translation of specific mRNAs required for synaptic function. The product of the pumilio (pum) gene was shown to repress local eIF4E accumulation at synapses of the NMJs by binding directly to the mRNA of eIF4E. Local eIF4E levels were found to be relevant for the modulation of the translation efficiency of the glutamate receptor GluRIIA mRNA and thus for synaptic transmission [75]. Furthermore, overexpression of eIF4E can cause malignant transformation of cells, an effect that can be reversed by reducing eIF4E levels. Accordingly, elevated levels of eIF4E are observed in cancer cells, correlating with the severity of the disease [29].
Another critical parameter for the modulation of translational efficiency within cells is the stability of the cap binding complex-mRNA interaction. In vitro experiments using recombinant eIF4E, eIF4G and PABP indicate that cap binding complexes are stable and dissociate relatively slowly from mRNAs. However, in cap-binding assays using purified human proteins, the addition of eIF4B, a factor with helicase activity, can destabilize preassembled cap complexes, leading to an accelerated turnover of cap binding complex-mRNA interaction [76]. This suggests that in vivo as opposed to in vitro eIF4B and possibly other factors may alter the stability of the cap complex.
In conclusion, changes in the concentration of eIF4E apparently produce two distinct types of response, one relating to general translation, which is at best subtle, and the other on specific aspects of cellular function, which can be in several cases considerable. A possible explanation for this discrepancy is that a specific subset of mRNAs within cells has a special requirement with respect to eIF4E function and is thus particularly sensitive to alterations in the level of functional eIF4E. Examples of these mRNAs are the yeast CLN3 transcript [74], and the VPF [77], FGF-2 [78], CyclinD1 [15], ODC [79], and Pim-1 [80] mammalian mRNAs. The regulation of these transcripts via eIF4E has been suggested to involve two possible different mechanisms. The first mechanism might be based on subtle differences in the affinity of the cap-binding complex for different mRNAs. Although, as discussed above, only minor differences have been pointed out in the capability of the cap-binding complex to discriminate between different mRNA species, these differences could become critical in the presence of limiting amounts of eIF4E within cells. The second one could involve 5′-UTR secondary structures and the consequent differential dependency on the helicase activity of eIF4A by different mRNAs to bind the small ribosomal subunit. Thus the limitation in the assembly of a cap complex might limit the resolution of secondary structures within a given mRNA and hence affect its translation.
3.3 eIF4E and nuclear export of mRNAs
Another aspect of transcript specific regulation of translation is the dependence of select mRNAs on eIF4E for their nuclear export. Despite the fact that the eIF4E-dependent mRNA export is still poorly characterized, the human cyclinD1 message and other mRNAs have been shown to be exported via an eIF4E-dependent mechanism [81,82]. Thus, protein synthesis for some mRNAs might be sensitive to eIF4E levels independently of translational control. Interestingly, a mutant of eIF4E that cannot function in translation but can mediate cyclinD1 mRNA export, is capable of transforming cells as efficiently as the wild-type protein, suggesting that the oncogenic capacity of the cap-binding protein may be based on the control of mRNA export rather than on translational control [43]. These results would furthermore suggest that a pool of free eIF4E might function in the export of specific mRNAs without significantly competing with eIF4E, bound to eIF4G, for binding to the cap structure. Two human proteins have been identified as additional interactors of eIF4E, which might function as negative regulators of eIF4E-dependent nuclear export of specific mRNAs: the PML (promyelocitic leukaemia) and PRH (proline-rich homeodomain protein) proteins [43,82]. The interactions between these proteins and eIF4E require the dorsal Trp residue of eIF4E as do the interactions with eIF4G and the 4E-BPs. However, PML and PRH do not contain the conserved eIF4E binding motif, although PRH contains a related sequence in which the hydrophobic residue (Φ) is exchanged for a glutamine [82]. While 4E-BPs and eIF4G stabilize binding of eIF4E to the cap structure, both PML and PRH reduce the affinity of eIF4E for the cap structure. The negative effect on eIF4E's cap binding activity is probably linked to the regulatory role of PML and PRH on the export activity of eIF4E.
Interestingly, in addition to PRH and BCD other homeodomain transcription factors have been recently reported to be able to directly interact with eIF4E. Indeed, among the known homeodomain-containing proteins about 200 were found to contain a putative eIF4E binding sequence [82]. One of these, the HOXA9 homeodomain protein, was shown to functionally interact with eIF4E. HOXA9, however, unlike PRH, appears to be a stimulator of eIF4E activity. HOXA9 can antagonize the interaction of PRH with eIF4E thereby modulating the PRH-mediated repressive action on eIF4E activity. HOXA9 apparently modulates both the nuclear as well as the cytoplasmic functions of eIF4E. In the nucleus, HOXA9 was found to promote the eIF4E-dependent nuclear export of the cyclinD1 and ornitine decarboxylase (ODC) mRNAs, whereas in the cytoplasm it was found to stimulate the efficiency of translation of the ODC mRNA. [83]. Similarly, the Emx2 homeodomain transcription factor was found to be associated with eIF4E in the olfactory sensory neuron axons, suggesting that it may play a role in controlling the local translation of mRNAs encoding proteins mediating synaptic plasticity and/or axon guidance [84] (Table 1). It seems likely that additional homeodomain protein–eIF4E interactions will be reported in the future reinforcing the knowledge that control of translation via eIF4E may be directly linked to the activity of factors controlling patterning and regional identities in development.
3.4 eIF4E and mRNA turnover
Several pathways of mRNA degradation require removal of the cap structure since the presence of the cap inhibits the activity of exonucleases that are crucial for this process (Fig. 2). Specific decapping enzymes have been found in several organisms; the best characterized are the yeast Dcp1 and Dcp2 (reviewed in [85]). Dcp cleavage activity on the cap requires access by these enzymes to parts of the cap structure that are engaged in eIF4E binding. Evidence both in vitro [86,87] and in vivo [86,88] for a competition between eIF4E and Dcp activity have been provided. It can be thus speculated that a decrease in eIF4E and eIF4G activity could lead to mRNA degradation, and more generally cap-binding complex destabilization could correspond to an acceleration of mRNA degradation. In yeast, the shortening of the poly(A) tail to about ten nucleotides is a prerequisite for decapping and subsequent mRNA degradation. Since the interaction of the yeast poly(A) binding protein (Pab1) with the poly(A) tail was shown to require a larger number of nucleotides [89], it could be hypothesized that poly(A) tail shortening could lead to PABP dissociation and the consequent destabilization of the eIF4G–eIF4E-cap interaction, rendering the cap accessible for Dcp activity. However, yeast strains that carry a mutation in Pab1, able to bind poly(A) but not eIF4G, show normal deadenylation and decapping activities [90], indicating that the correlation between poly(A) tail length and mRNA turnover in cells is less obvious than predicted. Removal of the cap structure requires in addition the activity of a number of accessory factors such as the enhancers of decapping Edc1 and Edc2, the mRNA-binding protein Pat1, the Lsm1–7 proteins, which bind Dcp proteins and are recruited to the mRNA after deadenylation, and the RNA helicase Dhh1 [91–93]. These proteins were shown to associate with mRNAs forming a large complex after deadenylation but before decapping [93]. Thus rather than by a direct competition between Dcps and eIF4E, mRNA stability could be influenced by the competition for cap access between the aforementioned decapping accessory factors and eIF4E. According to this view, shortening of the poly(A) tail would induce association of the Lsm1–7 complex to the mRNA, leading to the dissociation of the cap-binding complex via the helicase activity of Dhh1 and the consequent gain of access of Dcps to the cap [93].

Schematic representation of eukaryotic mRNA turnover. Several pathways of mRNA degradation require removal of the cap structure since the presence of the cap inhibits exonuclease activities that are crucial for this process. The first step is the shortening of the poly(A) tail to about ten nucleotides, a prerequisite for removal of cap structure by Dcp enzymes. Following decapping, mRNA is degraded by 5′–3′ exonucleases. Another pathway of mRNA degradation independent of decapping is not represented here.
3.5 eIF4E phosphorylation
In addition to its association with inhibitory eIF4E-binding proteins, eIF4E is regulated through phosphorylation [16]. The MAP kinase-interacting kinases 1 and 2 (Mnk1 and Mnk2), which are phosphorylated and activated by the mitogen-activated protein kinases (MAPKs) ERK and p38, phosphorylate eIF4E on Ser209 [94], the major site of phosphorylation of eIF4E in mammals [95–97]. In mice lacking both the Mnk1 and Mnk2 genes, eIF4E is indeed not detectably phosphorylated at Ser209 [98]. However, these mice are viable, fertile, develop normally, and show neither general protein synthesis nor cap-dependent translation defects, suggesting that eIF4E phosphorylation at the conserved Ser209 is not essential for cell growth during development [98]. Surprisingly, Mnk1 does not interact with eIF4E directly, but binds to the C-terminal region of eIF4G. Thus, it has been proposed that eIF4G provides a docking site to bring Mnk1 next to its substrate eIF4E [99]. While Mnk1 appears to be the major physiological eIF4E kinase [16], protein kinase C (PKC) has been shown to phosphorylate eIF4E as well [97], and a PKC consensus sequence does indeed surround Ser-209 [96].
The phosphorylated form of eIF4E binds to mRNA caps 3–4-fold more tightly than the non-phosphorylated form [37]. However, recent findings have demonstrated that phosphorylation of eIF4E impairs its ability to bind capped mRNA [40,100] and increases the rate of dissociation of eIF4E from the cap analogues or capped RNA [40], suggesting that phosphorylation occurs only after mRNA binding. Thus it is a dephosphorylated eIF4E that binds an mRNA cap, leading to the formation of the translation initiation complex and recruitment of the ribosome.
Scheper and Proud [101] proposed two possible mechanisms related to eIF4E phosphorylation: (A) phosphorylation occurs immediately after the assembly of the initiation complex and facilitates eIF4E release from the cap structure, allowing the ribosome to begin scanning; (B) phosphorylation occurs later in the initiation process and enhances the release of initiation factors from the cap-structure, rendering the cap-binding factors available for the translation of other mRNAs. Thus, phosphorylation could play an important role in ‘reprogramming’ of the translational machinery.
Phosphorylation of eIF4E is enhanced by a variety of agents that stimulate translation, including insulin, hormones, growth factors, and mitogens [95,96,102]. Thus, it has been proposed that eIF4E phosphorylation plays a positive role in cell growth by enhancing translation rate [16]. Recently, genetic data in Drosophila demonstrated that eIF4E phosphorylation is biologically significant and necessary for normal growth and development, because a point mutation of the known eIF4E phosphorylation site (Ser 251, corresponding to Ser 209 of mammalian eIF4E) causes a delay in development and reduction in body size [19].
In Drosophila, the Lk6 kinase, the closest homologue of mammalian Mnk kinases, is responsible for eIF4E phosphorylation [103]. Loss of lk6 function leads to slower development, and reduced viability and adult size, demonstrating that Lk6 function is required for organism growth. Moreover, in lk6 mutant flies eIF4E phosphorylation is dramatically reduced [104]. Double Mnk1/Mnk2 knockout mice have demonstrated that both kinases are dispensable for development and growth. These results differ from the one obtained in Drosophila probably because either redundancies or compensatory effects occur in mammals [104]. In addition other factors may influence the phosphorylation status of eIF4E. The translational regulator Cup, which interacts with eIF4E to modulate development and growth of the Drosophila ovary, likely plays a role in the control of the phosphorylation status of eIF4E within the developing ovary, since in cup mutant ovaries the amount of phosphorylated eIF4E appears to be reduced [56].
In the marine mollusc Aplysia californica, eIF4E is phosphorylated at Ser207 (corresponding to Ser 209 of mammalian eIF4E) by protein kinase C [105]. An antibody that specifically recognizes the phosphorylated form of Aplysia eIF4E showed that the level of phosphorylated eIF4E correlates with the basal rate of translation in the nervous system [106]. Moreover, in this organism eIF4E dephosphorylation can trigger a switch to IRES-mediated translation [107]. eIF4E phosphorylation might be involved in synaptic plasticity and memory as well. In murine hippocampal neurons, reduction of ERK activity led to a decrease of neuronal activity-induced translation and phosphorylation of eIF4E and other translation factors [108].
4 CPEB and cytoplasmic poly(A) addition
The initial polyadenylation of an mRNA molecule occurs in the nucleus. Whereas the 7-methylguanosine cap structure is added at the 5′ end as soon as transcription has started, the 3′ poly(A) tail is added by the poly(A) polymerase (PAP) after transcription has ended. Nuclear poly(A) addition requires a series of cis-acting sequences and specific trans-acting proteins bound to them (Fig. 3). The nuclear polyadenylation signal AAUAAA, bound by the cleavage and polyadenylation specificity factor (CPSF), is positioned 10–30 nucleotides upstream of the polyadenylation site (represented by a 5′-CA-3′ di-nucleotide) that is followed, after 10–20 nucleotides, by a G/U-rich region bound by the cleavage stimulation factor (CstF). After binding of these factors, the emerging RNA molecule is cleaved, just downstream of the CA sequence, and the poly(A) polymerase adds 100–250 adenosine residues, according to the animal species, to the 3′ end [109]. These modifications confer mRNA stability, promote translation efficiency, and have a role in the transport of processed mRNA from the nucleus to the cytoplasm [110].
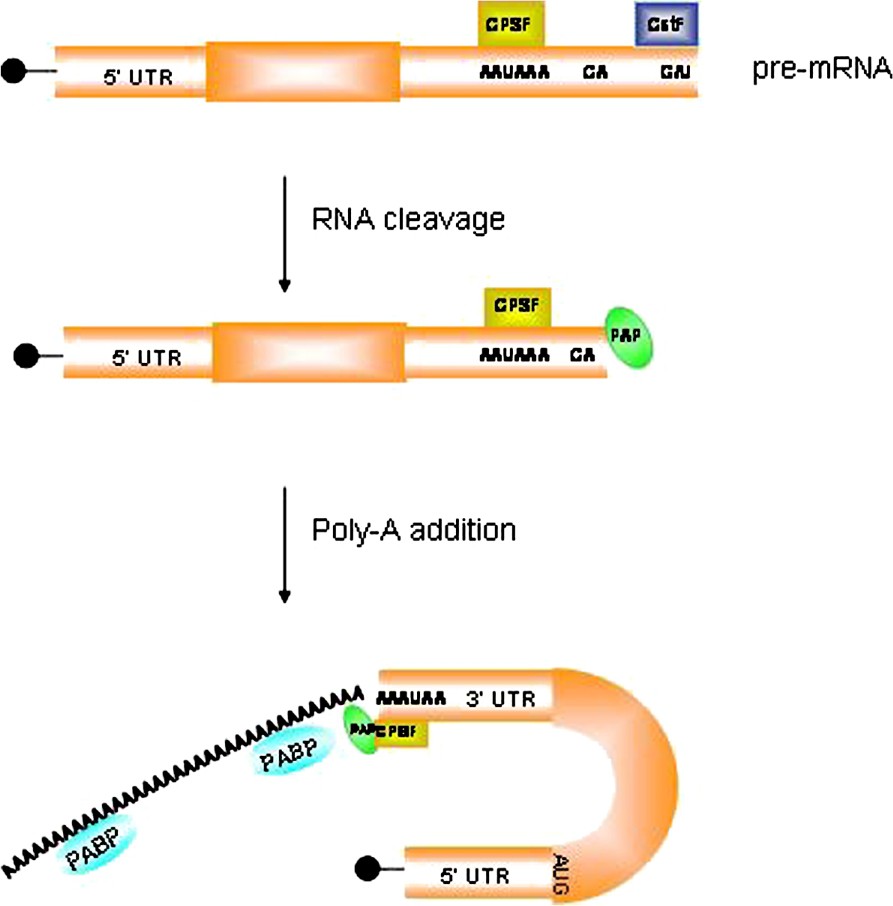
Mechanism of polyadenylation in the nucleus. The poly(A) tail is added by the poly(A) polymerase (PAP) after transcription has ended. Nuclear poly(A) addition requires a series of cis-acting sequences and specific trans-acting proteins bound to them. The nuclear polyadenylation signal AAUAAA, bound by the cleavage and polyadenylation specificity factor (CPSF), is positioned 10–30 nucleotides upstream of the polyadenylation site (represented by a 5′-CA-3′ di-nucleotide) that is followed, after 10–20 nucleotides, by a G/U-rich region bound by the cleavage stimulation factor (CstF). After binding of these factors, the emerging RNA molecule is cleaved, just downstream of the CA sequence, and the poly(A) polymerase adds 100–250 adenosine residues to the 3′end.
Once mRNAs emerge from the nucleus, the destiny of their poly(A) tails can vary: they are slowly shortened in most cells [111] thus gradually diminishing their translation rate, or undergo dramatic changes thus regulating translation at specific times and places in early development. During oocyte maturation and early embryogenesis, specific mRNAs destined to be stored in a translationally dormant state are deadenylated rapidly, causing their repression [112]. Later, the same mRNAs receive a long poly(A) tail and become translationally active [111]. Oocytes and early embryos are transcriptionally inactive and require rapid changes in the proteins they contain to regulate development [111]. A form of translation regulation is the poly(A) tail lengthening of specific mRNAs, a process called cytoplasmic polyadenylation and driven by cytoplasmic enzymes.
4.1 Translational regulation through cytoplasmic polyadenylation
Cytoplasmic polyadenylation is generally correlated with translational activation and deadenylation with translational repression. Translational control by cytoplasmic polyadenylation is necessary for mouse and Xenopus oocyte maturation as well as Drosophila embryogenesis [111,113–115]. Induction of translation of many maternal mRNAs, such as c-mos and tissue plasminogen activator (tPA) in vertebrates [112,113,115], and bicoid, Toll, torso, and hunchback in Drosophila [116,117] is accompanied by their cytoplasmic polyadenylation and plays an essential role in the early development of these organisms. The cytoplasmic polyadenylation reaction, at least in Xenopus and mouse, is regulated by sequences in the 3′ UTR of mRNA that include the hexanucleotide AAUAAA (a highly conserved sequence required also for nuclear pre-mRNA cleavage and polyadenylation) and U-rich sequences (e.g., UUUUUAU) called cytoplasmic polyadenylation elements (CPEs). CPEs activity can be modulated also by other sequences located in the 3′-UTR [118–120]. Insertion of a CPE into the 3′UTR of a reporter mRNA causes polyadenylation and translational induction during oocyte maturation or early development [112,118,119,121]. However, not all mRNAs that contain CPEs undergo cytoplasmic polyadenylation, suggesting that this sequence alone is not sufficient to induce polyadenylation [114]. At least three factors are involved in cytoplasmic polyadenylation: the poly(A) polymerase, similar to nuclear poly(A) polymerases in mammalian cells [122], CPSF [123], also known to be essential in nuclear polyadenylation, and CPE-binding protein (CPEB).
CPEB, a 62-kDa protein with two RNA recognition motifs (RRMs) [124] and a zinc finger domain [125], was first described as a protein critical for cytoplasmic polyadenlytion in Xenopus oocytes. Immunodepletion of the protein from Xenopus egg extracts renders them incapable of in vitro polyadenylation, while partial polyadenylation activity is restored by supplementing the depleted extract with in vitro synthesized CPEB [124]. CPEB is an RNA-binding protein that binds specifically to the CPEs and controls the polyadenylation of several mRNAs, including those encoding c-Mos, several cyclins, and cdk2 during oocyte maturation in Xenopus. In Xenopus, c-mos mRNA encodes for a serine/threonine kinase which regulates oocyte maturation [126]. Removal of two cis-acting sequences contained within the 3′-UTR of endogenous c-mos mRNA results in the complete inhibition of oocyte maturation [115]. Polyadenylation of c-mos mRNA is also required for its translational activation in mice and this process is dependent on the presence of two CPEs in the 3′-UTR [113]. Injection of CPEB antibody into oocytes blocks their progesterone-induced maturation due to inhibition of cytoplasmic polyadenylation and translation of c-mos mRNA, thus suggesting that CPEB is critical for early development [127]. Male and female CPEB gene null mice are viable and develop normally, but are infertile [128]. In addition, oocytes of CPEB knock-out animals fail to polyadenylate and translate CPE-containing mRNAs [128].
The initiation of cytoplasmic polyadenylation requires the activity of the Eg2 kinase, a member of Aurora family of serine/threonine protein kinases. CPEB is phosphorylated on Ser174 in vitro, and immunodepletion of Eg2 from oocyte extracts prevents CPEB phosphorylation on this amino acid residue. CPEB phosphorylation is both necessary and sufficient to stimulate polyadenylation and translation of c-mos mRNA [129]. Moreover, phosphorylation of CPEB increases its affinity for CPSF [129], which in turn interacts to the AAUAAA sequence and recruits poly(A) polymerase to the 3′ end of the mRNA (Fig. 1B) [123,130].
The mechanism of polyadenylation-induced translation is still unclear; it has been proposed that in response to elongation of the poly(A) tail some mRNAs undergo a 5′ cap-specific 2′-O-methylation that enhances translation efficiency during oocyte maturation [131]. A second mechanism may involve the poly(A) binding protein (PABP). Long poly(A) tails are bound efficiently, every 20–30 residues, by PABP that is able to bind also eIF4G. This has led to the idea that the association of PABP with mRNAs bearing long poly(A) tails can help to recruit eIF4G for binding with eIF4E thus promoting translation [93,132]. PABP contributes to translation in Xenopus oocytes and a mutant form of eIF4G, unable to interact with PABP, reduces translation of polyadenylated mRNAs and inhibits maturation of oocytes [133].
CPEB has been described as a protein able to activate translationally dormant mRNAs in Xenopus oocytes by elongation of their poly(A) tails [24]. CPEB, however, does not act only in activation of translation, since it has also been shown to repress translation of mRNAs bearing a CPE in the 3′-UTR [134]. Thus, CPEB, which appears to be constantly bound to specific cis-acting sequences in the 3′-UTR of mRNA, mediates both repression and activation of translation and these opposing behaviours may be explained by its diverse interactions. The repressive role of CPEB involves the interaction with Maskin, which was isolated as a CPEB-binding protein. Maskin contains an eIF4E-binding motif [16,52] and bridges eIF4E with CPEB thus preventing the binding of eIF4G, required to correctly position the 43S ribosomal subunit on the mRNA, to eIF4E and blocking initiation of translation of CPE-containing mRNAs (Fig. 1D) [135]. The Maskin-eIF4E complex is disrupted by cytoplasmic polyadenylation triggered by CPEB phosphorylation [136], allowing eIF4G to bind eIF4E and activate translation [52]. Translation repression is relieved when PABP binds to the poly(A) tail and helps eIF4G displace Maskin and bind eIF4E (Fig. 1B) [52].
4.2 CPEB, CPEB-like molecules, and transcript localization during early development
CPEB homologues have been identified in humans (hCPEB) [137], clams (p82) [138], flies (Orb) [139] and zebrafish (Zorba) [140]. In addition, four isoforms have been cloned in mice (mCPEB-1, mCPEB-2, mCPEB-3, mCPEB-4) [141,142] and four CPEB homologues have been identified in Caenorhabditis elegans (CPB-1, CPB-2, CPB-3, and FOG-1) [143,144]. In worms, two CPEB homologues, CPB-1 and FOG-1, have key functions in spermatogenesis and are dispensable for oogenesis [144], whereas in frogs, flies, and clams, CPEBs are essential during oogenesis [120,139,145] (Table 2).
CPEB-like molecules and their functions in development
| mRNA targets | Function in development | |
| xCPEB (Xenopus laevis) | c-mos | Oocyte maturation |
| Cdk2 | Oocyte maturation | |
| cyclin B1 | Oocyte maturation | |
| p82 (Spisula solidissima) | cyclin A | Oocyte maturation |
| Orb (D. melanogaster) | oskar | Posterior determinant |
| mCPEB (Mus musculus) | c-mos | Oogenesis/Spermatogenesis |
| CPB-1 (C. elegans) | N/A | Spermatogenesis |
| FOG-1 (C. elegans) | N/A | Spermatogenesis |
| hCPEB (H. sapiens) | N/A | N/A |
In Drosophila, translational activation of bicoid (bcd) mRNA is required for determination of anterior structures in the embryo. The poly(A) tail of the bcd mRNA is extended after fertilization and bcd is translated at that time. A long poly(A) tail is required and might be sufficient for translation of bcd [116]. For certain mRNAs, however, the extension process, instead of poly(A) tail length per se, seems to be necessary to activate translation [119,146]. In Drosophila, no cis-acting elements involved in cytoplasmic polyadenylation have been identified so far. However, CPEB is 62% identical to Orb [124], an oocyte-specific RNA-binding protein involved in RNA localization and in antero-posterior and dorso-ventral patterning during Drosophila oogenesis [139,147]. The loss of orb activity in Drosophila oocytes blocks the polyadenylation and translation of certain maternal mRNAs [139,147,148]. The Drosophila CPEB homolog Orb binds directly to oskar mRNA 3′UTR and activates oskar translation [148]. mRNA localization appears mediated by sequences located in the 3′UTR of localized transcripts [149]. Multiple cis-acting elements within the 3′-UTR of orb appear indeed to be required for directing its own localization during Drosophila oogenesis [139].
Localization-dependent translation is a common translational regulatory mechanism in development [150]. In this context, mRNAs can be localized at specific sites within the cell where they are translated. oskar mRNA is localized to the posterior pole of the oocyte where Oskar protein, a posterior determinant, directs posterior cell fates [151,152]. Translation of oskar mRNA is repressed before the localization process commences. An ovarian protein, named Bruno, has been implicated in this translational repression. The repressor protein Bruno binds specifically to sequences called BRE (Bruno Response Elements) present in the 3′UTR of oskar mRNA [153]. Moreover, it has been shown that a germ-line protein, named Cup (see eIF4E section), acts together with Bruno to repress oskar translation (Fig. 1F) [53]. Bruno shares a 50% sequence identity with the Xenopus deadenylation promoting factor EDEN-BP [154], EDEN-dependent deadenylation promotes rapid poly(A) tail shortening of a set of maternal mRNAs after fertilization of Xenopus oocytes and represents likely a conserved mechanism to inhibit translation in metazoa [154].
Posterior localization of oskar mRNA is essential for its translation. oskar mRNA is transported to the posterior pole of the oocyte, where Orb protein is already localized, by the RNA-binding protein Staufen [155,156]. Orb then activates translation of osk by inducing its cytoplasmic polyadenylation, since oskar mRNA in orb mutant flies has a shorter poly(A) tail than in wild type [148]. Orb mutants localize oskar transcripts with a shortened poly(A) tail that fails to enhance oskar translation, thus resulting in posterior patterning defects [157]. In contrast, it has been proposed that translation of oskar mRNA could be poly(A) independent [158,159] leading to the idea that polyadenylation is not the determining event of translation derepression. Cytoplasmic polyadenylation might be important for enhanced and efficient translation [157].
A growing body of experimental evidence demonstrates that CPEB is expressed also in neurons [160,161]. The protein translation machinery is present in dendritic processes and it has been proposed that specific mRNAs may be translationally repressed before they reach the dendrites where repression is relieved [161,162]. CPEB, controlling translation of specific mRNAs located in dendritic processes, may promote synaptic plasticity within dendrites [161,162]. CPEB may be activated following synaptic stimulation and promote the polyadenylation and translation of CPE-containing mRNAs in dendrites [58,161,163]. CPEB facilitates mRNA translation as well as transport [161,163]. CPEB also colocalizes with Maskin in CPE-containing RNA particles that are transported along microtubules to dendrites [163]. Overexpression of CPEB enhances RNA transport, whereas overexpression of a CPEB mutant protein, which is unable to associate with kinesin and dynein, inhibits transport [163]. In Aplysia it has been identified a neuron-specific isoform of CPEB that regulates the synaptic protein synthesis in an activity-dependent manner [164].
4.3 CPE- and CPEB-independent cytoplasmic polyadenylation
The mechanism of translational regulation by cytoplasmic polyadenylation appears to be tightly coupled to the binding of CPEB to CPE sequences. In progesterone-stimulated Xenopus oocytes, however, early cytoplasmic polyadenylation, and subsequent translational activation, of a class of maternal mRNAs occurs independently of a CPE and CPEB [165]. On the contrary, late cytoplasmic polyadenylation occurs only via CPE- and CPEB-dependent mechanisms. A novel type of sequences contained within the 3′-UTR of early mRNAs, named polyadenylation response elements (PREs), temporally direct early cytoplasmic polyadenylation and translational activation. This element was originally identified in the 3′-UTR of Mos mRNA and shown to function during Xenopus oocyte maturation [166].
5 Conclusion and future challenges
Multiprotein complexes regulate translation initiation of eukaryotic mRNAs by modulating the cross-talk between the cap structure and the poly (A) tail. The 5′ and 3′ termini of mRNAs represent therefore major targets for translational control during development and pathological conditions such as cancer.
The binding of eIF4E to the cap structure is modulated by its phosphorylation. It is not yet clear, however, whether phosphorylation increases or diminishes the binding affinity to the 7-methyl-guanosine and remains to be ascertained if multiple phosphorylation sites are involved in this process. Genetic and biochemical data indeed suggest that phosphorylation of the evolutionary conserved Ser209 (human coordinates) alone does not explain all its effects in vivo. Proteomic techniques coupled to genetic analysis, obtained in a simple animal model system such as Drosophila, could unravel the modality and role of phosphorylation during eukaryotic translation initiation and early development.
CPEB-dependent translational regulation promotes germ-line development in diverse organisms, varying from flies to mammals. Mental deficits in humans are often linked to sterility, thus suggesting a role of CPEB in learning and memory activities. Moreover, the existence of multiple CPEB isoforms in mammals may imply tissue-specific translational regulatory mechanisms. A multi-organisms approach will help unravel the inhibitory/stimulatory mechanisms of CPEB and its multiple targets.
In recent years data from many laboratories have uncovered the general mechanisms of protein synthesis. Novel mRNA-specific translational regulatory mechanisms continue however to be discovered and the number of factors involved in such regulations is increasing proportionally. In addition, several distinct eIF4E isoforms, with specific developmental expression profiles, have been identified in Drosophila [167] and a similar scenario could be soon unravelled for members of the CPEB family. It is therefore expected that more examples of regulative protein interactions will be reported in the near future, providing support to the idea that eIF4E and CPEB functions can be regulated by the interaction with a wide variety of tissue and/or body region-specific proteins in different contexts. This information will contribute to clarify the molecular links underlying the essential steps towards both translation repression and translation activation of mRNAs in different organs at various time in development.
Acknowledgements
We apologize to researchers we have not cited due to space limitations. Work in the authors' laboratories is supported by grants from the Italian Ministry of Education (MIUR).


