1 Introduction
Polyploidy is a major evolutionary process, which is considered to have promoted speciation, contributed in the adaptation of organisms to new ecological niches, or helped the acquisition of novelties to face environmental changes [1–5]. In plants, numerous examples of polyploids have been reported and it is clearly demonstrated that ancestors of all present species have experienced several rounds of polyploidisation. Variable polyploidy states have also been reported in some species, even at the population level (e.g. [6,7]). In long-lived species such as trees, spontaneous intraspecific ploidy variation has been commonly observed, for instance, in poplar [8], oak [9], ash [10], elm [11], or acacia [12], but frequency of polyploids remains usually low in natural tree populations (e.g. less than 1% in Quercus robur [9]).
Polyploidy was recently evidenced in two relict wild subspecies of the olive tree (Olea europaea L., Oleaceae) in South Morocco (subsp. maroccana [Greut. & Burd.] P. Vargas et al.; [13]) and Madeira (subsp. cerasiformis G. Kunkel & Sunding; [13,14]). It was proposed that such polyploids were advantaged in very small endangered populations by limiting effects of inbreeding depression. However, the origins of these polyploids remain poorly understood and deserve some investigations [15]. In particular, the mechanisms involved in the formation of such polyploids are unclear.
Previous studies also demonstrated that the polyploidy level of olive trees can be inferred from nuclear microsatellite data offering an alternative to the fastidious chromosome counting; subspp. maroccana and cerasiformis were thus suspected to be respectively hexaploid and tetraploid from genetic analyses, and this was then confirmed by flow cytometry analyses [13,14]. Some intriguing genetic profiles were also reported in the Laperrine's olive [O. europaea subsp. laperrinei (Batt. & Trab.) Ciferri] populations, since a few trees with three different alleles at several microsatellite loci were observed in the Hoggar and Air mountains [16–19]. The diploid status of these trees was thus questioned but not more investigated, since no living material was available in collection for a flow cytometry analysis.
In the present study, we report the characterisation of a population of Laperrine's olive with eight variable nuclear microsatellite loci coupled to a flow cytometry analysis to determine the ploidy of a few trees displaying a complex genetic profile. A statistical approach was also developed to assess the efficiency of a microsatellite approach for the detection of triploid genotypes.
2 Material and methods
2.1 Study species and sampling
The populations of O. e. laperrinei have regressed with the desert expansion during the Pleistocene [20] and are generally constituted by small patches distributed in high mountains from central Sahara to northeast Sudan. This taxon persists in very dry areas particularly in southern Algeria and northern Niger where mean rainfall can be less than 50 mm per year [21]. Today, the regeneration of the Niger and Algerian O. e. laperrinei populations is considered as null, probably due to very long drought periods and an intensive browsing by wild and domesticated animals [16,20]. As a consequence, clonal growth is favoured and allowed the maintenance of very old individuals [16–19].
In the present study, a population of Laperrine's olive from the Adrar Heggueghene massif in the Hoggar (southern Algeria) was considered. In this massif, several hundreds of olive individuals are distributed in a fragmented habitat on cliffs and borders of temporary rivers. Leaf samples of 106 genets were collected on five distinct sites during several field campaigns (Table 1). For each sample, DNA was extracted from a single leaf using a CTAB method [22].
Geographic origin of the plant material characterised in the present study. The number of diploid (2x) and triploid (3x) genets is given for each site.
| Site | Altitude | Longitude | Latitude | 2x⁎ | 3x⁎ |
| Tahâssa | 1480 m | 22°47 N | 5°39 E | 14 | 0 |
| I-n-Toûnine | 1390 m | 22°45 N | 5°38 E | 6 | 1 |
| I-n-Ezzebib | 1470 m | 22°48 N | 5°37 E | 22 | 0 |
| Hadriane | 1600 m | 22°47 N | 5°36 E | 23 | 1 |
| Ti-n-Hâmor | 1600 m | 22°51 N | 5°37 E | 38 | 1 |
| Total | 103 | 3 |
⁎ Ploidy level as deduced from microsatellite profiles (see Supplementary material).
2.2 Genotyping and data analysis
Most of the sampled individuals were partially characterised in previous studies with different sets of nuclear microsatellite loci [16,19] but a few new samples from Hadriane (15 individuals) were added in the present study. Genotyping of all these trees was completed for eight microsatellite loci (DCA01, DCA03, DCA05, DCA08, DCA09, DCA15, DCA18, GAPU71A [23,24]) according to the methodology described by Baali-Cherif and Besnard [16]. We only considered loci with very clear genetic profiles (i.e. no ambiguity for allele size estimation), and some previously used loci, such as DCA14 and EMO-03 that display 1-bp variation (due to polymorphism in mono-nucleotide stretches), were excluded. The complete microsatellite genetic dataset is available in the online Supplementary material.
The number of alleles (), allelic frequencies (), and Nei's gene diversity () were computed for each locus L with Fstat [25] from 103 individuals displaying diploid genotypes (see below). The probability for a triploid genotype at locus L, , to display three distinct alleles i, j and k was computed as:
The probability of detecting a triploid genotype at least at one locus was thus given by:
2.3 Cytometry analyses
Cuttings of two genets (“I-n-Toûnine 1-2-3”, “Hadriane 19”; see below) displaying three different alleles at three or four loci, and therefore suspected to be triploid, were collected for a flow cytometry analysis. The genet “Hadriane 19” is presently maintained in a living collection at the INRA Montpellier. Two additional Laperrine's olive trees from Ti-n-Hâmor (“Ti-n-Hâmor 1” and “6”) with one or two different alleles at all loci were also analysed as diploid references. The 2C-value (nuclear DNA content) of these trees was estimated using flow cytometry as reported by Besnard et al. [13]. The measures were done three times independently for each individual.
3 Results and discussion
3.1 Evidence of triploids in the Laperrine's olive population
As expected from previous studies [16–19], the eight loci used to characterise the Laperrine's olive population are polymorphic, and five of them display relatively high number of alleles () and/or gene diversity (; Table 2). Among the 106 genets analysed, we found three individuals (2.8%) with three different alleles at three or four loci (i.e. “I-n-Toûnine 1-2-3”, “Ti-n-Hâmor 9-10” and “Hadriane 19”; Supplementary data; Fig. 1). These individuals were sampled on three relatively distant sites. The remaining 103 trees display one or two different alleles at each locus. The diploid status of genets “I-n-Toûnine 1-2-3”, “Ti-n-Hâmor 9-10” and “Hadriane 19” can thus be questioned. The 2C-value for two of them (“I-n-Toûnine 1-2-3” and “Hadriane 19”) was therefore measured by flow cytometry and was respectively 4.97 pg (±0.05) and 4.91 pg (±0.02). A 2C-value of 3.34 pg (±0.17) was obtained for two diploid Laperrine's olive trees. This latter estimate is congruent with previous 2C-values reported in diploid olives from different geographic origins [13,14]. These observations clearly support that “I-n-Toûnine 1-2-3” and “Hadriane 19” have a genome about 1.5 bigger than diploid trees, and we can thus contend that they are triploid as also suggested by their microsatellite profiles [9].
Characteristics of the eight microsatellites used for the Laperrine's olive genotyping.
| Locus | Allele range | ⁎ | ⁎ | |
| DCA1 | 226–284 | 26 | 0.92 | 0.782 |
| DCA3 | 229–253 | 7 | 0.70 | 0.318 |
| DCA5 | 199–249 | 22 | 0.90 | 0.688 |
| DCA8 | 119–147 | 12 | 0.84 | 0.580 |
| DCA9 | 169–193 | 12 | 0.56 | 0.196 |
| DCA15 | 251–258 | 2 | 0.23 | 0.000 |
| DCA18 | 152–183 | 12 | 0.81 | 0.561 |
| GAPU71A | 217–225 | 4 | 0.63 | 0.215 |
| Total | – | 97 | 0.70 |
⁎ Measured on the 103 diploid profiles.
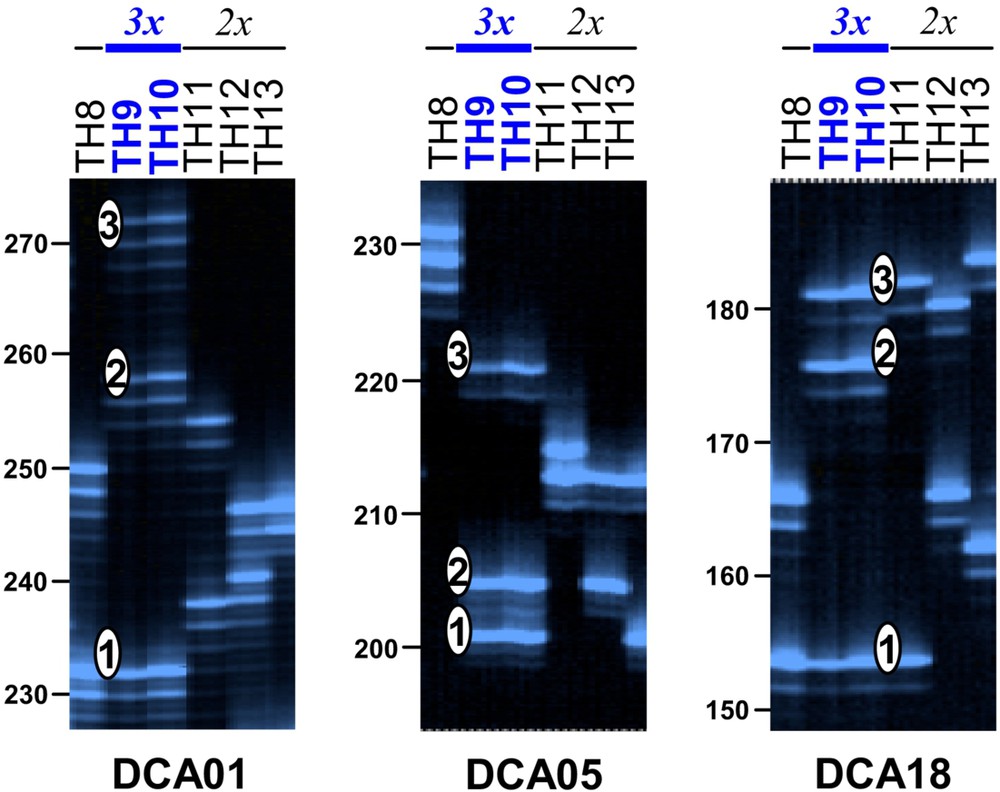
Examples of microsatellite profiles for diploid (TH8, TH11, TH12 and TH13) and triploid (TH9-10) individuals from Ti-n-Hâmor at loci DCA01, DCA05 and DCA18. The three alleles observed in the genet TH9-10 (“Ti-n-Hâmor 9-10”) are numbered 1 to 3. Fragment-size scale (in bp) is indicated on the left of each picture.
The efficiency of our eight microsatellite loci to detect triploid genotypes is very variable (; Table 2). The two more efficient ones are DCA05 and DCA01. These loci display the highest gene diversity and number of alleles, and theoretically allow triploid detection in 69 and 78% of cases, respectively (Table 2). By using the seven loci with more than three alleles (excluding DCA15), 99.5% () of triploids are expected to be revealed. All of them have therefore been very likely detected in the studied population. However, we should also mention that the presence of only one locus with three different alleles should not be enough to conclude on the triploid status of a tree. Indeed, it is not excluded that mutations can lead to chimerism at some loci as demonstrated for instance in grapevine [26]. In the Laperrine's olive, somatic mutations have been clearly reported at nuclear microsatellites on different ramets issued from the same stump (e.g. [16,18]), but such chimeras were generally not observed in a single leaf (e.g. one case was previously suspected for genet “I-n-Toûnine 1-2-3” [16] but the presence of three alleles at locus DCA01 was finally due to triploidy as demonstrated in the present study). The maintenance of chimerism in a single organ first depends of the position of the mutation in a meristem. To produce a relatively stable chimera, a new mutation needs to be located on the apical dome of a meristem and this only concerns a small proportion of mutants (i.e. periclinal chimera [26]). However, the stability of such chimera can also be affected by diplontic selection favouring one cell lineage [27,28]. For the genotyping of individuals, we extracted DNA from a single leaf and we can thus expect that, for diploids, the frequency of loci with more than two different alleles (due to somatic mutations) should be very low. Therefore, detection of triploids based on evidence from several microsatellite loci (as in the present study) is expected to be very efficient.
3.2 On the origin of triploidy in the Laperrine's olive
Variable ploidy levels in a population of the same subspecies were never reported in O. europaea, and all previous microsatellite studies of the “diploid” Mediterranean and African olives did not reveal complex genetic profiles in cultivars (e.g. [29]) as well as in wild populations (e.g. [15,30–32]). This suggests that the pattern observed in the Laperrine's olive is quite unusual in the O. europaea complex, and this leads us to wonder on the significance of the triploidy in the Laperrine's olive. Some molecular evidences suggest that recurrent migrations from both Mediterranean and Tropical Africa contributed to establishment of the present Laperrine's olive populations [33]. In flowering plants, spontaneous polyploids have been shown to be more common in hybrid systems [1], and the intraspecific hybrid status of the Laperrine's olive could have thus been an important factor in the polyploidy formation. However, additional experiments based on single-copy genes are needed to test more accurately such hypotheses on autopolyploidy vs. segmental allopoyploidy. Several explanations can also be proposed to explain triploid formation. Some tetraploid individuals could have crossed with diploids to lead to triploids. Tetraploids have never yet been detected in the Laperrine's populations, but it is possible that somatic polyploidisation could have sporadically led to mixoploid individuals, and then the polyploidised shoot (i.e. ) could be involved in the production of triploids as already demonstrated in an olive mixoploid mutant [34]. The triploids could have also resulted of the zygote development from the endosperm, or more likely, should be from one gamete that fails to undergo meiotic reduction [1,9].
A relatively high proportion of triploids (2.8%) is observed in our olive population if we compare it to frequencies observed in different species of oaks (from 0.25 to 0.57% in natural populations [9]) or acacias (from 0 to 1.3% in seedling populations [11]). In two Laperrine's olive populations from North Niger, one individual with a typical polyploid microsatellite profile was also detected among 98 genotypes [18]. In contrast, we only observed diploids in 211 seeds/seedlings recently collected in the Hoggar Mountains and characterised with the same set of microsatellite loci [19] suggesting that triploid formation is sporadic. In plants, it has been reported that triploids are unusually vigorous when compared to diploids (e.g. [9,35]), probably because it reduces inbreeding depression effects, and this should favour their establishment in extreme conditions. During stages of sexual regeneration, inbreeding is expected to be quite high in the Laperrine's olive populations due to the distribution of trees in very small patches and limited gene dispersal [19]. Very long periods of vegetative growth without sexual regeneration should have allowed the maintenance of very adapted genotypes. In such conditions, the more vigorous trees should be favoured, and triploids are probably in mean more successful than diploids [36]. In a more extreme situation (e.g. strong isolation of a small population leading to high inbreeding), we can also speculate that this process could lead to the establishment of fully polyploid populations (e.g. 4x or 6x) as observed in Madeira and South Morocco, and the emergence of triploids in a diploid population could be interpreted as the first step towards such populations. Indeed, even if the fertility of triploids is usually considerably reduced when compared to diploids, several authors proposed that such genotypes might be involved in tetraploid or hexaploid formation (via a triploid-bridge [1]). This hypothesis should be experimentally tested by crossing genotypes with different ploidy levels (i.e. 2x and 3x) maintained in collection.
Supplementary material
Acknowledgements
We are grateful for the kind collaboration of Mohamed-Salah Adellaoui & Boudjemâa Azzouz, who helped us to collect the plant material for this study. We also thank Amélie Fragnière (Fribourg University) for her help in flow cytometry analyses and Céline Devaux for statistical advices. G.B. was funded by the Intra-European Fellowship PIEF-GA-2008-220813.


