1 Introduction
The soil is one of the major reservoirs of biological diversity on our planet [1]. Many processes and interactions take place in the soil, contributing to a considerable number of ecosystem services [2]. Pimentel et al. [3] estimated the economic profit derived from soil biodiversity at 1546 billion dollars, although the relative value of the associated services remains to be determined. Among the ecological, social and economic services identified, the role of soil as a reservoir of biodiversity has now been well established along with its role in surface water purification, the recycling of mineral elements (soil fertility), and carbon storage (as a sink for atmospheric CO2), this latter process being directly related to climatic changes and the notion of plant productivity [3]. However, studies involving quantification of soil biodiversity, and studies of its role in biological functioning are much less common than similar studies for aboveground organisms, in particular plants. Accordingly, our knowledge on the role of soil biodiversity remains limited. This is true for indigenous microbial communities which are still considered (on the basis of the postulate proposed by Beijerinck [4] that “everything is everywhere”), as ubiquitous communities exhibiting high functional redundancy. Telluric microbial communities are still repeatedly considered as a functional “black box” generating fluxes, the intensities of which are solely dependent on abiotic factors such as temperature, humidity, pH… This approach therefore excludes that the structure of microbial communities, as well as trophic interactions (competition, commensalism…) within these communities might play a functional role [5,6]. The resulting state-of-the-art is:
- • a lack of knowledge concerning the diversity of soil microorganisms, its spatial distribution at small- and large-scales, and its implication in soil functioning [7];
- • an insufficient, even absent, consideration of microbial diversity in the models currently used to quantify matter and energy fluxes [8].
Technical difficulties explain for a part these scientific lacunae in microbial ecology: difficulties in characterizing soil microbial diversity, as a result of the poor accessibility of indigenous microbial populations within the heterogeneous and structured soil matrix, and difficulties in deciphering information that represents 100,000 to 1,000,000 different species per gram of soil [9]. Another explanation comes from the non-consideration of spatial and temporal scales in microbial ecology, which precludes comparison and generalization of the obtained results. However, due to major advances in molecular biology during the past twenty years, techniques have been developed to investigate and decipher the diversity of soil microbial communities in situ and without a priori [10]. Moreover, various studies have been conducted at very large sampling scales to enhance the robustness of their conclusions [7].
In this scientific context, our short review aims at providing a better understanding of present and future prospects in soil microbial ecology. It is divided into three main sections. The first one is rather technical and constitutes a survey of the development of molecular tools, which makes it possible to investigate the diversity of soil microorganisms at a finer level. The second section emphasizes the need of integrating spatial upscaling into studies of soil microbial ecology to improve our knowledge of the large-scale determinism of microbial community assembly. The third and final section describes a methodological strategy that links soil biodiversity with ecosystem services.
2 Methodological developments to characterize soil microbial diversity: from Pasteur to soil metagenomics
The scientific domain described as “microbial ecology” is about 50 years old and is thus very young. Its step-by-step evolution has mainly been promoted by methodological developments [11]. In the 1960s, most comprehensive studies were focused on monoxenic cultures and little attention was given to possible interactions between microorganisms and their habitat. In the 1980s, one of the first advances was to consider not only single organisms but the density, diversity and activity of microbial populations isolated from natural environments. In the 1990s, this approach was used in many studies and provided the basis for our current understanding of the microbial world and its role in ecosystem functioning. At the same time, considerable effort was devoted to developing molecular methods to characterize the microbial diversity through the analysis of nucleic acids extracted from environmental samples. These developments permitted the characterization of variations in soil microbial community structure and diversity in a variety of situations and the identification of the populations preferentially associated with specific environmental perturbations [10,12]. These combined methodological developments led to medium-throughput genotyping and sequencing, thereby allowing analysis of the metagenome (total DNA from all microorganisms present in an ecosystem) and providing most of the microbial DNA sequences deposited in databases.
Since the development of these so-called “molecular ecology” approaches, the number of studies involving characterization of the microbial communities in various environments or subjected to different perturbations has increased exponentially [12]. All these studies are highly promising and, in some cases, are providing relevant insights into chronic or punctual modifications of microbial biodiversity in natural environments. However, they can often lead to contradictory results because the techniques and sampling strategies have not been standardized. For example, the estimated size of bacterial diversity in soils can be very different according to the technique used. It can reach 103 species per gram of soil for 16S rRNA PCR cloning and sequencing, 104 for 16S rRNA pyrosequencing and taxonomic microarray, and even 106–107 for reassociation kinetics of soil DNA [9,13,17]. Consequently, the results are difficult to compare and do not significantly increase our understanding of soil microbial communities, their biodiversity or their role in soil biological functioning.
2.1 Metagenomic strategy to assess soil microbial diversity
The recent development of high-throughput sequencing technology (pyrosequencing, Solexa, SOLiD) allows several tens of thousands, even hundreds of thousands of sequences to be obtained from a single genomic or metagenomic DNA, and shows promising potential for characterizing soil microbial communities [14,15] (Fig. 1).
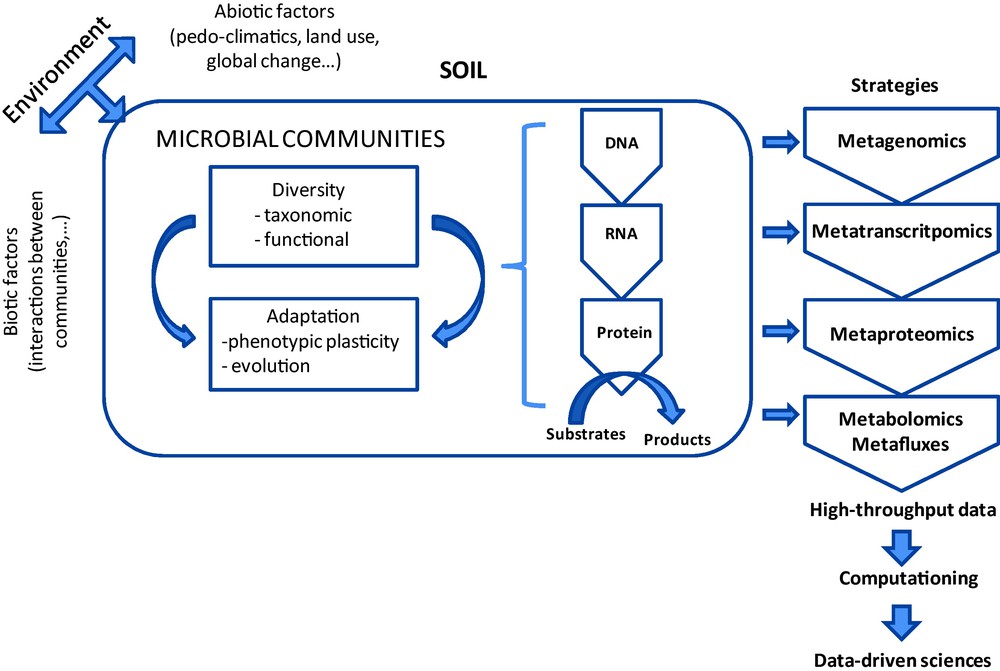
“OMIC” strategies to study soil microbial communities.
Metagenomic analysis should make it easier to decipher taxonomic and functional assemblages of indigenous communities in natural environments, determine their potential roles in the biological functioning of ecosystems, and identify the associated services. Two complementary strategies can be envisaged.
The aim of the first strategy is to analyze all the DNA sequences extracted from the indigenous communities in a given environment by targeting either a gene providing particular taxonomic information (ribosomic genes, corresponding to a “MetaTaxogenomic” analysis of microbial communities) or functional data (functional genes, corresponding to a “MetaFonctiogenomic” analysis of the microbial communities). This approach implies: (i) prior amplification of the target genes by PCR; (ii) the production of sequences that are large enough to be sensitive in terms of analysis of diversity (fine taxonomic level, ideally species); and (iii) the availability of a bioinformatics analysis pipeline to generate diversity indices and taxonomic affiliations. The acquired data can be used to process a large number of environmental samples (100 > N > 1000). However, only a few precise and exhaustive investigations of soil bacterial diversity, involving the use of pyrosequencing techniques, have been referenced to date. Two of them were carried out on four soils originating from distant geographical locations (USA, Canada and Brazil, [14]). Between 11,000 and 21,000 OTU/gram of soil were described for the 50,000 sequences analysed per soil. Such studies represent the first exhaustive descriptions of the enormous richness of soil bacterial diversity. More recently, Fulthorpe et al. [15] produced an alternative analysis of these data and demonstrated the weak similarity in community composition between the soils, thereby revealing that distantly-sampled soils carry few species in common. These data support the hypothesis that various pedoclimatic characteristics, as well as land use and soil management history, can lead to induce different indigenous microbial diversities.
Pyrosequencing techniques were also used in other investigations to decipher soil microbial diversity and elucidate the distribution of diversity of a particular taxonomic group of soil bacteria. Jones et al. [16] sampled and characterized soils from North to South America (from Alaska to Patagonia) to determine the influence of abiotic soil parameters on the abundance of Acidobacteria. They used this approach to define the ecological attributes of the targeted groups and to rank the environmental parameters that most explained their spatial distribution.
Limited knowledge of the taxonomic and functional sequences is one of the main blocks restricting our ability to identify new species or new functional genes. Although few studies have been published as yet, the scientific community is unanimous in affirming the relevance and enormous potential of this type of approach for characterizing the diversity of soil microorganisms [17]. Sampling on a huge spatial scale should make it possible to characterize this diversity, determine its spatial distribution and better understand the determinism of this distribution.
The second strategy is based on the mass sequencing of DNA fragments, cloned or not. This approach has proved particularly useful in analyzing metagenomes from the Sargasso Sea [18], an acid mine [19], the digestive tube [20] and soil [21]. The enormous quantity of genomic data needed to guarantee sufficient coverage of the diversity together with the tremendous scale of the associated processing and analyses will require huge financial and analytical inputs. These inputs will often exceed the national context and necessitate the setting up of international projects, which in turn will imply the creation of an international technical and scientific consortium [21]. This approach should make it possible to thoroughly explore the metagenomic diversity of a soil and also to look for new activities by applying high flow-rate screening methods for cloned sequences. Due to the considerable financial means that are required, only a limited number of situations can be analyzed (one soil at present). The data will then need to be subjected to bioinformatics analysis. This raises a certain number of questions as to the amount of data to be stored and processed which means, in turn, that the bioinformatics aspect is of pivotal importance in determining our capacity to process the information produced and make it accessible: a new bioinformatics strategy needs to be invented to process this huge quantity of data. At present, analyses of metagenomic data are based on clusters of sequences presenting certain levels of similarity. This clustering is then annotated by analogy with sequences in the databases. In the case of a cluster that cannot be assimilated with known information, an innovative approach defined as “meta-proteogenomic” and combining the metagenomic and metaproteomic approaches, has been proposed [22].
Both strategies have strengths and weaknesses, so progress in deciphering soil microbial communities and their agroenvironmental implications should be facilitated by their combined application. The first strategy allows the targeting of a limited number of genes and essentially provides access to the taxonomic and functional diversities of the communities involved in particular ecosystemic services [14]. In contrast, it allows a very large number of environmental situations to be examined and thus appreciation of a variety of environments. Studies based on the first strategy should increase our knowledge of diversity on large spatial scales and provide the necessary elements of information to propose “steering tools” and ultimately even to establish models to forecast the evolution of diversity and its consequences on soil biological function and the ecosystem services rendered. The second strategy provides a “complete” characterization of the diversity of a given soil and thus the cognitive information essential to our understanding of its biological functioning. These pilot projects are also essential to continue the development of sequencing technology and of bioinformatics analysis tools to analyze the metagenomes of other soils, given the expected evolution and cost-reduction of the sequencing techniques. Soil metagenomics and the strategies used for their study are the subject of debate both at national and world levels, as attested by the exchanges of correspondence via scientific publications [21,23,24].
The second strategy could be applied in efforts to sequence the entire genome of cultivated bacteria which harbor particular metabolic pathways or where an experimental approach could be helpful in annotating genes with unknown function, produced as a result of the metagenomic sequencing effort [20]. The metagenomic sequencing data would then be used to compile a soil gene catalogue.
3 Spatial distribution and determinism of soil microbial diversity: from small to large scale
3.1 Spatial scale: a major challenge in soil microbial ecology
Although these recent advances in molecular biology have produced tools for the assessment of microbial diversity in environmental samples without cultivation, most studies have focused on cataloguing the bacterial diversity in particular sites and describing how bacterial communities are affected by environmental perturbations. Nearly all studies of the diversification of prokaryotes have focused on variations due to mutations and/or lateral gene transfers and subsequent selection due to environmental stresses and competition for resources. Few studies have considered more neutral mechanisms, such as genetic drift brought about by spatial isolation and/or climatic variations, thus revealing the crucial lack of integration of the spatial scale into microbial community assembly [25,26]. As a result, the data obtained from different studies are difficult to compare and the trends deduced are often inconsistent. One relevant approach to enhance the general applicability of studies in soil microbial ecology is to investigate the “beta” and “gamma” diversity of soil microbial communities (i.e., changes in community composition on a large scale such as landscape and country, respectively). These large-scale investigations are further justified because they correspond to human activities (agriculture, industrialization, urbanization). However, characterizing microbial biodiversity require powerful and robust tools (see section 2) and application of these tools to enormous numbers of samples.
Studies of the abundance and diversity of microbial communities and populations on small and large scales have systematically revealed that their distribution is both heterogeneous and spatially-structured. This observation does not support the hypothesis that “everything is everywhere” deduced by Baas Becking [27] from the works of Beijerinck [4], which suggests that all microbial populations are cosmopolitan. On a microscale, the heterogeneous distribution of soil microbes is mainly determined by soil structure and porosity (governing water stress, aeration and predation) and by organic carbon content (governing trophic resources) [25]. On a field scale, the microbial diversity resulting from natural and/or anthropogenic modifications of surrounding perturbations has been examined in numerous studies. The main factors structuring community abundance and diversity are physicochemical characteristics, such as texture [28], pH [29] and organic status [30] of the soil, but also include soil management [31] and plant cover [32]. Although numerous investigations have led to more or less precise determination of the drivers of community diversity and composition on a field scale, the results obtained from different pedoclimatic zones cannot be generally applied or the drivers ranked according to their relative influence on soil microbial diversity.
3.2 Biogeographical patterns of invisible life
Ecologists studying plant and animals have long recognized that an examination of the modifications in diversity across a landscape is pivotal to understanding the environmental factors that drive the magnitude and variability of that diversity. However, this conceptual vision is also relevant to microorganisms since it can offer valuable insights into the relative influence of dispersal limitations, environmental heterogeneity, and environmental and evolutionary changes in shaping the structure of communities. Despite the statement that spatial patterning of microbial diversity can have important consequences on plant community structure and ecosystem functioning, few studies have been carried out on a wide spatial scale. The environmental filters structuring microbial biodiversity are still largely unknown. In this context, the empirical relationship between the number of species and area sampled (taxa–area relationship) has not been investigated in microorganisms, contrary to plants and animals [33,34].
Thus, there is a need to find out if microbial diversity is governed by the same laws as that of macroorganisms or if the particularities of the microbial component (minute size, rapid generation time, huge diversity) lead to specific patterns of distribution on large spatial scales. The question remains open since this aspect has been tackled in very few studies (partly because of the above-mentioned technical limitations and the difficulty in generalizing the obtained results).
Microbial ecologists describing biodiversity on a wide spatial scale (i.e. microbial biogeography) generally invoke one of the oldest fundamental paradigms in microbial ecology “everything is everywhere, but, the environment selects” [27]. The first part of this paradigm “everything is everywhere” is supported by several particularities of the microbial model: microorganisms: (i) are small and easily transported; (ii) have the ability to form a resistant physiological stage that allows them to survive in hostile environments; and (iii) form extremely large populations with a high probability of dispersal and a low probability of local extinction. More than 1018–1020 microorganisms are estimated to be transported annually through the atmosphere between continents, which supports the hypothesis that microbes are widely dispersed. Bacteria can also be isolated from places where “they should not be” such as thermophilic bacteria from cold seawater. On the contrary, the second part of the paradigm “the environment selects,” which challenges “everything is everywhere”, suggests that geographic population isolation coupled with limited dispersal leads to local and particular speciation.
To date, the studies dealing with the biogeography of the soil microbial community are few in number and insufficient to answer the different questions concerning the spatial distribution of microbes:
- • Are microbial communities a “black box” with no spatial structure or do they exhibit, like macroorganisms, a particular distribution with predictable, aggregated patterns on local to regional scales? In other words, does a taxa–area relationship exist in microbial-biogeography? [33,34].
- • Are spatial variations brought about by contemporary environmental factors or by land use history and contingencies? [35]
- • Which environmental factors (edaphic, climatic, land use, anthropogenic) most contribute to the structure and diversity of the soil bacterial community on wide geographic scales? [35]
Combined data from studies dealing with the biogeography of soil microbial communities have demonstrated:
- • a significant but moderate diversification of microbial communities on a large scale [33–35];
- • a hierarchy in the influence of environmental parameters with a strong influence of soil characteristics (notably pH) and also modes of soil use [36];
- • a weak influence of climatic and geomorphological parameters on composition of the communities and a total independence of the composition of such communities with regard to the geographical distance separating them [26].
It can be concluded from these results that the biogeography of microorganisms differs fundamentally from that of macroorganisms. Nevertheless, most studies of microbial biogeography have been based on non-systematic sampling and stratified with a priori concerning soil type or land use. This means that it is impossible to define large-scale profiles of geographical distribution and still less to compile maps of the distribution of microbial diversity. In this context, a scientific program (ECOMIC-RMQS) based on systematic sampling over the entire French territory has been developed to assess the biogeographical patterns of soil microbial communities.
3.3 A French initiative: the ECOMIC-RMQS project
The “ECOMIC-RMQS” project was set up in 2007, in the above-described scientific context, to offer the first opportunity to investigate biological diversity within a pre-existing large-scale soil monitoring system. The aim of this program was to characterize the density and diversity of microbial communities in the French “Réseau de Mesures de la Qualité des Sols = RMQS” which is a soil sampling network based on a 16 × 16 km systematic grid covering the whole of France [37]. The RMQS includes 2150 monitoring sites, each one located at the centre of a 16 × 16 km cell. Each site has been geopositioned with a precision < 0.5 m and detailed descriptions obtained of the soil profile, site environment, climatic factors, vegetation and land-use.
In this program we have shown that:
- • spatial distribution of the abundance and genetic structure of the bacterial communities is heterogeneous but structured into biogeographical profiles [38];
- • local parameters (soil type, land use), have a greater influence than global parameters (climate, geomorphology), on the distribution and therefore determinism, of the abundance and diversity of telluric bacterial communities [38,39];
- • diversification of the bacterial communities on the scale of a geographical region is closely linked to the diversity of the landscape (pedoclimate and usage) in that region (Ranjard et al., personal data);
- • certain modes of land use (notably agricultural) may have deleterious effects on the abundance and genetic structure of the communities [38,39].
This program was able to demonstrate the interest and feasibility of using the soil survey network to characterize soil microbial communities on a large scale and to address both fundamental (microbial biogeography, spatial ecology) and operational questions (environmental evaluation of land use). In addition, the program demonstrates that “putting microorganisms on a map” provides a relevant way to understand how environmental filters determine community assembly.
4 Functional interest of soil microbial diversity
At present, and regardless of its major involvement in ecosystem processes, the microbial component of soil is not taken into account in mathematical models designed to predict the fate of major elements in the environment [8]. Indigenous microbial communities are still considered in these models as a “black box” with a very high level of functional redundancy. As a result, only the size of the microbiota (biomass), but not its diversity or the composition of this pool, are integrated into such models. This is mainly due to a lack of knowledge of the microbial populations involved in soil functioning, and particularly of the contribution of microbial diversity in ecosystem processes. Until recently, two main reasons were put forward to justify this pragmatic approach:
- • these “black box” models are operational from a conceptual point of view even if microbial diversity is not taken into consideration;
- • understanding of the diversity of the microbial communities involved in ecosystem processes has been restricted, mainly because of the methodological limitations mentioned in section II.
Thanks to progress in modeling and new methods in molecular microbial ecology (see section 3), these reasons are no longer justified. Recent work on modeling the priming effect (i.e. the increase in soil organic C mineralization following the input of a fresh organic C compound) suggests that the density of microbial communities, together with competition between different functional groups in the microorganisms, may control the rate of mineralization of native soil organic matter [40–44]. These theoretical studies have highlighted the possibility that microbial diversity plays a functional role in organic matter turnover in soil and clearly suggest that this parameter should be included in the models. They have also evidenced the need for an empirical demonstration of the functional role of microbial diversity in ecosystem processes.
As mentioned in section 3, regarding the spatial distribution of biodiversity, the question of a link between diversity and ecosystem functioning, which is fundamental to functional ecology, has been addressed mainly by ecologists studying macroorganisms, particularly plants, through experiments involving the manipulation of taxonomic diversity or that of functional groups [45]. The first studies were reported in 1843, and concerned experiments set up on the English experimental station at Rothamsted [46]. This long history explains the abundance of literature currently available. The various investigations have produced a rich theoretical and conceptual framework, which has greatly improved our understanding of how biodiversity can influence ecosystem functioning [47]. Despite sometimes contradictory results, it is clear that plant diversity has positive effects on the functioning, performance and stability of ecosystems, and thus on their capacity to provide ecosystem services [48,49]. However, the question remains with regard to microorganisms since very few studies have examined this aspect in depth [45,50].
The literature available [45,51,52] indicates that simplification is essential to an experimental demonstration of the link between microbial diversity and ecosystem processes. Manipulating microbial diversity in controlled experimental systems appears to be a promising approach in this context, since the effects of environmental factors can be minimized and focus concentrated on the effects of microbial diversity. This strategy was first adopted in investigations by Salonius in 1981 [53]. It has subsequently been implemented in several studies to establish the relationship between microbial diversity and functioning in different environments [52,54–60]. In particular, these works revealed the importance of functional redundancy in governing the stability and resilience of the activities of soil microbial communities after a perturbation [51,54]. Experiments performed on the grassland systems of Jasper Ridge (California) by Horz et al. [56] showed that a modification in the diversity of the soil nitrifying community, brought about by global changes (increased atmospheric CO2, temperature, N deposition and soil moisture), could increase nitrification, suggesting that microbial diversity might play a pivotal role in the nitrogen cycle. In contrast, studies to date of the involvement of microbial diversity in the carbon cycle have not provided clear evidence to support such a link [58,60], although some studies have suggested it [61–63]. One reason may be that, within the soil microbial community, the decomposition of organic matter in soil is a highly redundant function. However, analysis of the enzymatic capacities required for soil organic matter degradation has highlighted the non-uniform distribution of these capacities within the soil microbial component. In particular, those involved in the final degradation steps are carried by only a small subset of the soil microbial community [64,65]. This could explain the successions of microbial populations observed during the degradation of plant residues added to soil [31,66,67] and suggests that microbial diversity may have a functional role in the decomposition of soil organic matter. In agreement with this hypothesis, Bell et al. [52], and Liebich et al. [55] showed that microbial diversity may play a role in the degradation of plant residues. However, both studies used different consortia of microbial species that had previously been isolated on culture media. Since only 1 to 10% of soil microorganisms can be cultured, this strategy is hampered by a strong selective bias and the resulting microbial consortium cannot be considered as representative of the indigenous communities [51]. In addition, the species richness of the reconstituted communities (10 species, Liebich et al., [55]; and 72 species Bell et al., [52]) was completely unrealistic with regard to the huge genetic diversity that characterizes the soil microbial world [68]. Consequently, the role of microbial diversity in the carbon cycle in soil requires further investigation and new fundamental studies will have to be carried out.
In contrast to earlier investigations of this topic, future studies will be able to take advantage of the recently developed DNA Stable-Isotope Probing (DNA-SIP) method that allows specific characterization of the communities actively involved in the decomposition of C-substrates labeled with stable isotopes (e.g., 13C) [66,69–72]. Another advantage of using labeled compounds is the possibility to monitor not only mineralization of the labeled C substrate, but also that of native soil organic matter (SOM) at the same time as the dynamics of the degrading communities responsible for the decomposition of each C-pool. It thus provides an opportunity to evaluate the importance of microbial diversity in ecosystem processes such as the priming effect (i.e. the increased mineralization of native SOM following the addition of fresh organic matter to the soil) that may play an important role in soil carbon balance [41,66,73,74]. Use of such a method in experiments, including the manipulation of microbial diversity, may constitute a decisive step towards experimental demonstration of the functional significance of microbial diversity in C-cycling in soil.
Nevertheless, extrapolation of the conclusions derived from investigations performed under simplified controlled conditions, will obviously be limited and results will need to be generally applicable in order to acquire a truly predictive dimension. A promising complementary strategy to achieve this goal in future will be to combine the powerful and robust tools used to characterize microbial biodiversity (i.e.: pyrosequencing of 16S rDNA gene from soil samples) and in functional ecology, with extensive sampling on a massive spatial scale (landscape, region, country or continent). With such a strategy, it should be possible to establish: (i) a statistical link between the different parameters of soil microbial diversity (i.e. synthetic diversity index/species richness/evenness) and intensity of the microbial processes involved in the major biogeochemical cycles, this being a prerequisite to determining the importance of microbial diversity in the provision of ecosystem services by the soil environment; and (ii) the relative importance of the environmental filters controlling the functionality of soil microbial communities. Large-scale sampling, as in national soil survey networks (for example the RMQS), offers unique opportunities to implement this strategy [75]. The final goal will be to propose large-scale maps (territory, region, landscape…) of the capacity of soils to perform ecosystem processes/services as a function of microbial diversity.
Conflict of interest statement
No conflict of interest.
Acknowledgments
The ECOMIC-RMQS project is granted by ADEME (French Environment and Energy Management Agency) and by the French National Research Agency (program ANR-06-BDIV-011). RMQS soil sampling and physico-chemical analyses were supported by a French Scientific Group of Interest on soils: the “GIS Sol”, involving the French Ministry for Ecology and Sustainable Development (MEDAD), the French Ministry of Agriculture (MAP), the French Institute for Environment (IFEN), the Environment and Energy Management Agency (ADEME), the French Institute for Research and Development (IRD) and the National Institute for Agronomic Research (INRA). We thank all the soil surveyors and technical assistants involved in sampling the sites. The link between soil microbial diversity and functioning is currently investigated in the DIMIMOS project granted by the ANR Systerra (2008–2012).


