1 Introduction
Aethionema R. Br. (Brassicaceae) is a taxonomically complex genus, and a few macromorphological characters are available for species delamination. Aethionema is a genus of flowering plants, within the family Brassicaceae, subfamily Brassicoideae. They originate from limestone mountainsides in Europe and Western Asia, especially Turkey. Life duration (annular/perennial) and fruit morphology are of impotence at the species level in Aethionema. The genus has its center in Turkey, and outside Anatolia its population declines very rapidly [1]. The Turkish flora is comprised of about 41 Aethionema species, of which 20 species are endemic to Turkey [2–4].
Most species of the Brassicaceae develop fruits in which seeds are released through a process termed fruit dehiscence. Some genera in Brassicaceae develop indehiscent fruits that do not release ripe seeds. The genus Aethionema is the sister group to all other extant Brassiceceae, some species of Aethionema are heterocarpic, meaning that they develop as both dehiscent and indehiscent.
Earlier classifications and evaluations of the genus Aethionema were done primarily based on phenotypic expressions of the plants such as growth form, leaf morphology, fruit properties such as color, length of the styles in female flowers and other agronomical characters, but information from these environmentally influenced morphological and physiological characteristics are not sufficient to identify Aethionema because the differences between them are often subtle and misleading. Hence, recent advances in the field of molecular biology and gene technology have been successfully used for the evaluation of the genetic relationship between plants species in addition to their morphological characters [5–10]. The RAPD and ISSR techniques, when compared to AFLP and SSR, are fast and easy, since they do not require knowledge of the sequences of the markers and can produce abundant polymorphic fragments. So far, there have been few attempts to study the genetic variation in Aethionema species using molecular analyses [11,12]. However, no studies have been conducted to assess the genotypic differences in most of Aethionema species, including those encountered in Eastern Anatolia (Turkey). Therefore, this study aimed at determining the genetic relationship among Aethionema caespitosum, Aethionema arabicum, Aethionema cordatum, Aethionema fimnraitum, Aethionema armenum, Aethionema speciosum supsp. speciosum, Aethionema memraneceum, Aethionema grandiflorum var. grandiflorum species growing in Eastern Anatolia using RAPD and ISSR technologies. The information obtained will be useful in the genetic analysis of Aethionema species.
2 Materials and methods
2.1 Plant material
A sample collection of eight species of Aethionema (A. caespitosum, A. arabicum, A. cordatum, A. fimnraitum, A. armenum, A. speciosum supsp. speciosum, A. memraneceum, A. grandiflorum var. grandiflorum) was collected at the flowering stage from different locations in the vicinity of Erzurum, Bayburt, located in Eastern Anatolia, Turkey (Table 1). Plant materials were ground in a grinder equipped with a 2-mm diameter mesh. The powdered plant material was then used for DNA extraction. The voucher specimen has been deposited at the herbarium, Department of Biology, Atatürk University, Erzurum, Turkey (Table 1). Plants were collected around Erzurum in 2011–2012 and deposited at ATA (Atatürk University Herbarium).
Aethionema species tested in this study.
| Aethionema species tested in this study | Herbarium number of the Vouchers | Locality | Status | Altitude |
| Aethionema caespitosum (Boiss.) Boiss. | 9848 | Kop mountain | End | 2100 |
| Aethionema arabicum (L.) Andrz. Ex DC | 9849 | Erzurum | — | 2250 |
| Aethionema cordatum (Desf.) | 9850 | Kop mountain | — | 2180 |
| Aethionema fimnraitum Boiss. | 9851 | Erzurum Hınıs |
Ir–Tur | 2200 |
| Aethionema armenum Boiss. | 9852 | Erzurum Hınıs |
Ir–Tur | 2350 |
| Aethionema speciosum Boiss. & A. Huet | 9853 | Kop mountain | Ir–Tur | 2200 |
| Aethionema memraneceum (Desu.) DC | 9854 | Erzurum Hınıs |
— | 2300 |
| Aethionema grandiflorum Boiss. & Halenvar. grandiflorum | 9855 | Erzurum Tekman |
— | 2100 |
2.2 DNA extraction
Genomic DNA was extracted from powdered plant materials using a method described by Sunar et al. [13]. The purity and quantity of genomic DNA was determined spectrophotometrically and confirmed using 0.8% agarose gel electrophoresis against known concentrations of unrestricted lambda DNA.
2.3 RAPD amplification
Forty-five primers had been attempted to generate RAPD profiles. Fourteen of these primers were selected: they produced amplicons with all of the Aethionema species tested, which were used in further studies based on the results of the preliminary tests (Table 2). PCR amplification reactions were carried out in a final reaction mixture volume of 30 μl, containing 10 × Buffer 3.0 μl, dNTPs (10 mM) 1.2 μl, magnesium chloride (25 mM) 1.2 μl, primer (5 μM) 2.0 μl, Taq DNA polymerase (5unit) 0.4 μl, water 19.2 μl sample DNA 3.0 μl (100 ng/μl). The thermal cycler (Eppendorf Company) was programmed as 2 min at 95 °C; 2 cycles of 30 s at 95 °C, 1 min at 37 °C, 2 min at 72 °C; 2 cycles of 30 s at 95 °C, 1 min at 35 °C, 2 min at 72 °C; 41 cycles of 30 s at 94 °C, 1 min at 35 °C, 2 min at 72 °C; followed by a final 5-min extension at 72 °C, then brought down to 4 °C.
Details of banding pattern revealed through RAPD and ISSR primers (R = A, G; Y = C, T).
| Markers | Primer/primer combination | Sequence (5′–3′) | Length of amplified bands | No. of bands | No. of polymorphic bands | Polymorphism ratio (%) |
| RAPD | A-1 | AGTCAGCCAC | 400–1800 | 7 | 6 | 85.7 |
| B-20 | GGACCCTTAC | 750–2200 | 8 | 8 | 100 | |
| C-10 | TGTCTGGGTC | 300–2000 | 5 | 5 | 100 | |
| OPBA-03 | GTGCGAGAAC | 500–2500 | 11 | 10 | 90.9 | |
| OPBB-03 | TCACGTGGCT | 500–1600 | 9 | 8 | 88.8 | |
| OPA-4 | AATCGGGCTG | 400–2300 | 8 | 8 | 100 | |
| OPA-13 | CAGCACCCAC | 600–2000 | 6 | 5 | 83.3 | |
| OPK-04 | CCGCCCAAAC | 750–2400 | 9 | 7 | 77.7 | |
| OPC-02 | GTGAGGCGTC | 250–2500 | 7 | 7 | 100 | |
| OPK-19 | CACAGGCGGA | 400–2100 | 12 | 12 | 100 | |
| OPL-15 | AAGAGAGGGG | 500–2600 | 6 | 6 | 100 | |
| OPY-13 | GGGTCTCGGT | 300–2500 | 10 | 9 | 90 | |
| OPD-20 | ACCCGGTCAC | 400–2800 | 7 | 7 | 100 | |
| OPH-10 | CCTACGTCAG | 600–2300 | 9 | 8 | 88.8 | |
| Total | 250–2800 | 113 | 105 | 92.9 | ||
| ISSR | UBC810 | (GA8)T | 400–2000 | 10 | 9 | 91.66 |
| UBC842 | (GA)8YG | 500–2400 | 7 | 7 | 100 | |
| UBC868 | (GAA) | 300–1800 | 8 | 7 | 87.5 | |
| UBC818 | (CA)8G | 600–3000 | 12 | 10 | 83.3 | |
| UBC825 | (CA)8T | 500–2600 | 8 | 8 | 100 | |
| UBC808 | (AG)8C | 400–2500 | 6 | 5 | 83.3 | |
| UBC811 | (GA)8C | 750–2000 | 7 | 7 | 100 | |
| Total | 300–3000 | 58 | 53 | 91.4 |
2.4 ISSR amplification
A total of 34 ISSR primers were tested for DNA amplification. Seven primers were chosen for ISSR analyses of genetic diversity, based on band reproducibly (Table 2). PCR reactions were carried out using a single primer at a time, in a 25-mL reaction mixture containing 40 ng of template DNA, 1 × reaction buffer, 200 mM of each of the four dNTPs, 1 U of Taq DNA polymerase, 1.5 mM of magnesium chloride and 0.5 mM of primer. Amplification was performed using a thermal cycler programmed for an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 1 min at the specific annealing temperature and 1 min at 72 °C, ending with a final extension step of 7 min at 72 °C. The PCR products of ISSR markers were resolved by electrophoresis on 1.5% agarose gels.
2.5 Electrophoresis
The PCR products (27 μl) were mixed with a 6 × gel loading buffer (3 μl) and submitted to agarose (1.5% w/v) gel electrophoresis in a 0.5XTBE (Tris-Borate- EDTA) buffer at 70 V for 150 min. The gel was stained in an ethidium bromide solution (2 μl EtBr/100 ml 1 × TBE buffer) for 40 min and visualized under UV in Bio Doc Image Analysis System with Uvisoft analysis package (Cambridge, UK).
2.6 Data analysis
PCR products were scored as presence (1) and absence (0) of band for each genotype and analyzed. Data were used to calculate a Jaccard (1908) similarity index, from which a dendrogram was constructed on the basis of this matrix by the UPGMA (unweighted pair group method using arithmetic average) method. Statistical analyses were performed using SPSS. All of the experiments in this study were repeated at least twice.
3 Results
Forty-five RAPD primers were initially screened against Aethionema species. Fourteen primers could produce a total of 113 distinct reproducible bands with an average of 8.1 bands per primer. The sizes of the amplified products ranged from 250 to 2800 bp. Of the 113 bands obtained, 105 (92.9%) were polymorphic. The number of polymorphic bands detected with each primer ranged from 5 (primer C10) to 12 (primer OPK-19).
A dendrogram constructed according to RAPD data (Fig. 1) of Aethionema species allowed us to divide them into two main clusters. The first cluster included A. caespitosum, A. cordatum, A. speciosum supsp. speciosum, A. armenum. The second one included A. arabicum, A. fimnraitum, A. memraneceum, A. grandiflorum var. grandiflorum. The greatest similarity was observed between species A. fimnraitum and A. memraneceum (0.185), the greatest dissimilarity was observed between species A. caespitosum and A. fimnraitum (0.948).
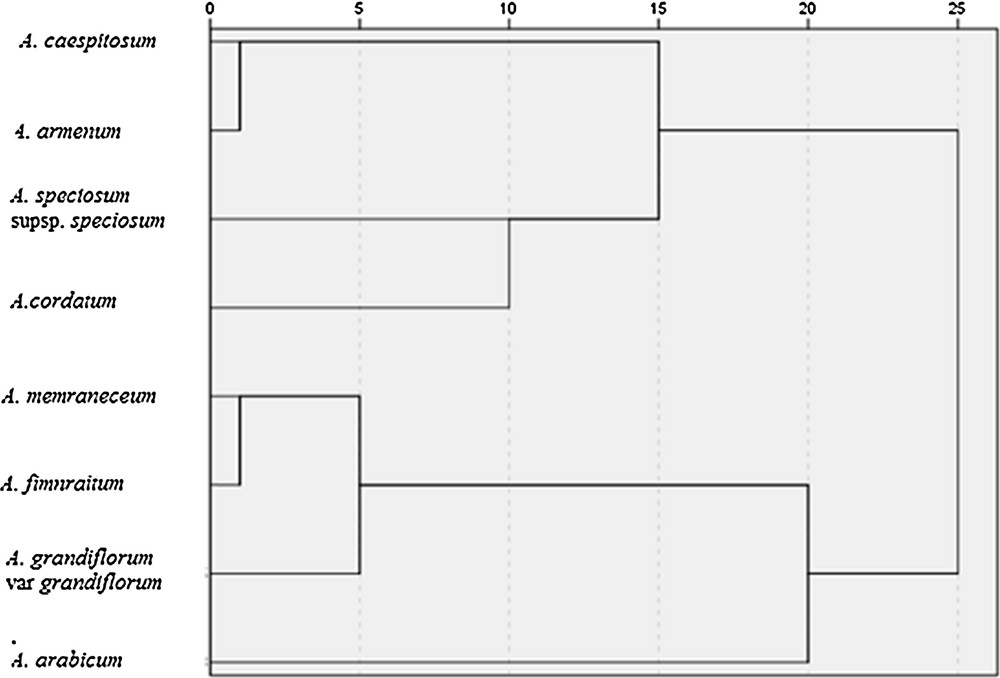
RAPD marker-based UPGMA clustering for eight Aethionema species.
Seven ISSR primers showing reproducible and polymorphic patterns were chosen for cultivar identification and generated a total of 58 bands, with an average of 8.3 bands per primer. The size ranged from 300 to 3000 bp. Of the 58 bands produced, 53 (91.4%) were polymorphic.
The number of polymorphic bands detected with each primer ranged from5 (primer UBC808) to 10(primer UBC818). A dendrogram that was constructed according to ISSR data of seven Aethionema species allowed us to divide them into two main clusters (Fig. 2). The first cluster included A. memraneceum, A. fimnraitum, A. armenum, A. grandiflorum var. grandiflorum, A. arabicum. The second cluster included A. cordatum, A. speciosum supsp. speciosum, A. caespitosum. The greatest similarity was observed between species A. memraneceum and A. fimnraitum (0.216), and the greatest dissimilarity was observed between species A. memraneceum and A. caespitosum (0.931).
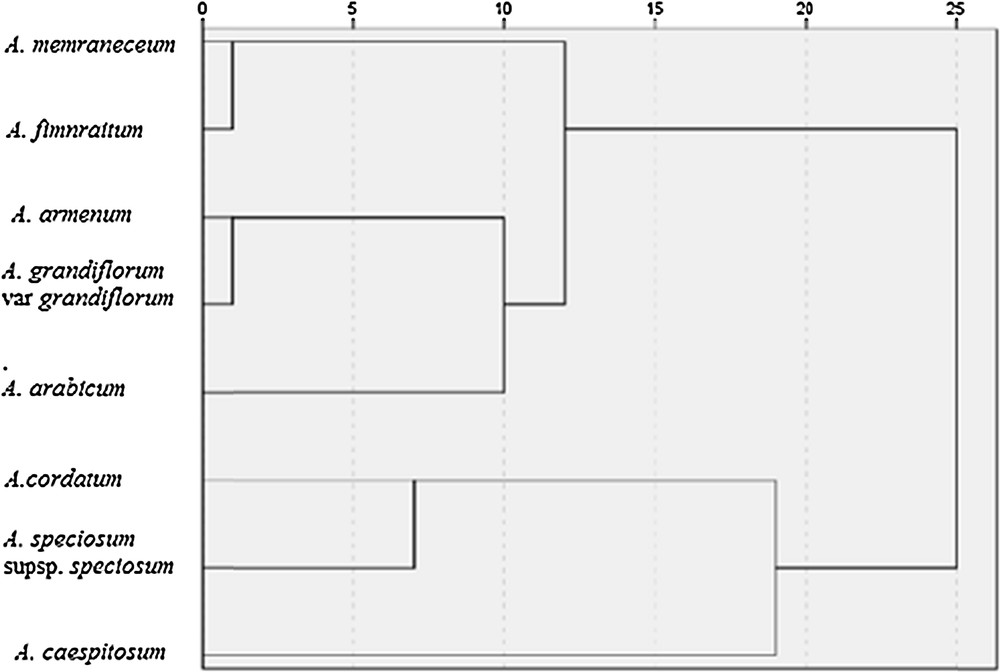
ISSR marker-based UPGMA clustering for eight Aethionema species.
The combined data generated from RAPD and ISSR marker analyses a dendrogram was constructed (Fig. 3). Eight Aethionema species were separated into two distinct clusters, each one with two species. The first cluster included A. cordatum, A. speciosum supsp. speciosum, A. caespitosum, A. arabicum. The second cluster included A. fimnraitum, A. memraneceum, A. grandiflorum var. grandiflorum, A. armenum. The combined analysis revealed that similarities of the species ranged from 0.182 (A. cordatum, A. speciosum supsp. speciosum) to 0.927 (A. grandiflorum var. grandiflorum, A. cordatum).
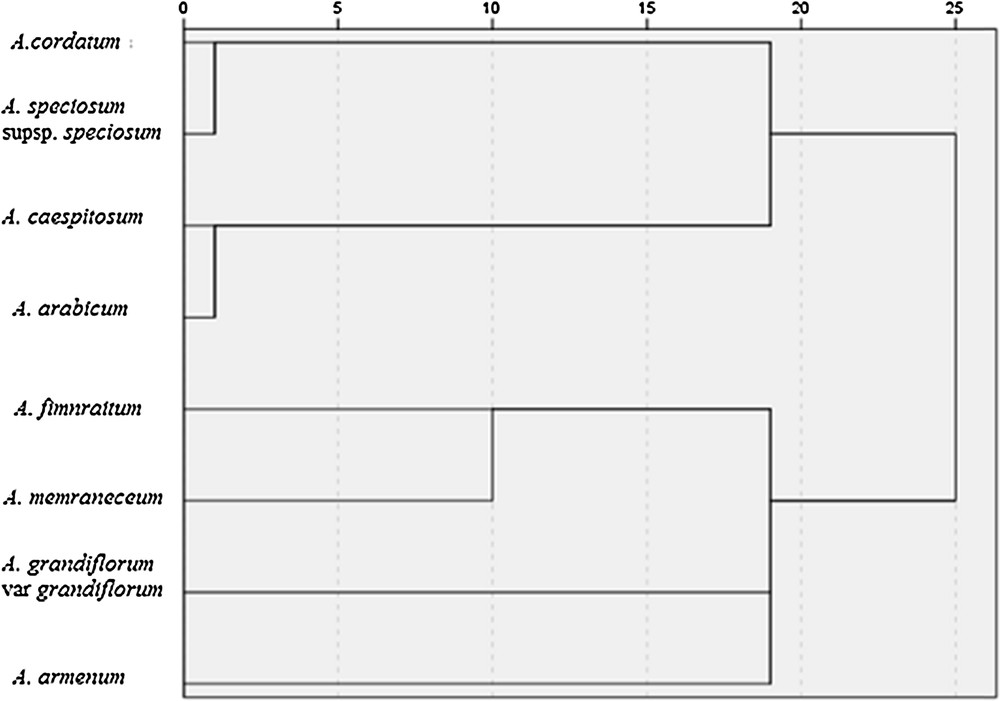
RAPD and ISSR marker-based UPGMA clustering for eight Aethionema species.
4 Discussion
The species of Aethionema tested in this study showed the presence of two clusters. Each cluster is represented by a different species of Aethionema, except for one cluster, which is divided into two subclusters and includes A. caespitosum, A. cordatum, A. speciosum supsp. speciosum, A. armenum and A. arabicum, A. fimnraitum, A. memraneceum, A. grandiflorum var. grandiflorum, which were the most genetically related species.
Both ISSR and RAPD profiles have been used to study phenotypic and genetic diversity in many plant species [11]. There have been a few attempts to study the genetic variations among the Aethionema species using molecular analysis [12,14], but none with the species tested in this study, except A. arabicum, A. grandiflorum.
Earlier studies using nuclear ribosomal DNA sequences (rDNA ITS) and ribosomal protein S 16 in the chloroplast genome (rps16) techniques showed genetic variations among A. carneum, A. semanensis, A. umbellatum, A. arabicum Andrz. Ex DC., A. elongatum Boiss., A. grandiflorum Boiss. & Hohen., A. saxatile R. Br., species. Khosravi et al. [12] reported little genetic variations among these species. However, molecular markers such as RAPD, ISSR, AFLP, SSR show considerable differences among species and genotypes. The RAPD and ISSR techniques when compared to AFLP and SSR are fast and easy, since they do not require knowledge of the sequences of the markers and can produce abundant polymorphic fragments Thus, RAPD- and ISSR-based molecular markers were able to distinguish between different species. This paper is the first report indicating the relationships between Aethionema species growing in Eastern Anatolia using RAPD and ISSR techniques. In conclusion, the results demonstrated that RAPD and ISSR analyses are useful for the differentiation of the Aethionema species tested in the present study.
However, additional phylogenetic studies using chloroplast or mitochondrial gene sequences or appropriate nuclear genes like ITS of nrDNA sequences can be helpful to reevaluate the systematic positions of these species.


