1 Introduction
Flax (Linum usitatissimum L.) is one of the oldest domesticated crops [1] with many applications. The long bast fiber of the species produces one of the oldest textile fabrics with extraordinary comfort due to unique mechanical properties and chemical composition [2,3], while short fibres together with wooden straw provide raw material for automobile, construction, package and paper industry (making cigarettes, currency notes, artwork) [4,5]. Flax seed is valued for its health-promoting properties due to the presence of bioactive agents like omega-3 fatty acids, lignans, mucilage [6–10] and is increasingly used in human food products [11,12]. Further, seed oil of the species is traditionally used in a range of industrial applications such as linoleum, paint, varnish, soap, printer ink, etc. To capitalise the expanding horizons of flax seed oil applications in the food industry, a number of low linolenic acid-containing varieties were developed [13,14]. Even seed cake, a byproduct of oil extraction used as a nutritious feed for livestock due to protein richness.
Cultivated flax along with another 199 species belongs to the genus Linum (Linaceae) [15,16] and has a chromosome number 2n = 30 [17]. The self-pollinating species is supposed to originate in either the Middle East or Indian regions, possibly from L. angustifolium (L. bienne) [17–20]. Molecular marker based studies comprehensively established its monophylogenic origin with oil type as a predecessor of fibre-type flax [21,22]. The oldest archaeological evidence for the wild species came from Tell Abu Hureyra in Northern Syria (11,200–10,500 years ago) [23] and by the eighth millennium BC the species was found distributed throughout the Near East [1]. The first cultivated form of flax archeologically was traced at Tell Ramad in Syria 9000 years ago and spread to Europe as well as the Nile valley as winter oil seed. However, in Eastern Europe, summer fibre-type flax evolved and gradually spread to central Europe [17,24]. On the other hand, the species was introduced into Western Europe (the Netherlands, Northern France, Belgium and Switzerland) between 5000 and 3000 BC by semi-nomads of the Middle East [25]. The available literature is silent about whether the domestication processes of fibre type flax at eastern Europe and middle East were independent events or not. Arguably, it is suggested that all modern fibre varieties in use today originated from Eastern Europe [24]. From there, the species travelled to America along with colonial labourers. During the trans-continental domestication process, the species evolved into different morphotypes depending on man-made differential preference either for fibre or seed oil. The century-long directed selection pressure led to the evolution of distinct morphotypes, particularly in the western and eastern regions of Eurasia with dominance of fibre and oilseed type, respectively [26]. Both morphotypes belong to the same species with distinct morphological, anatomical, physiological, and agronomical characteristics [17]. Presently Canada, India, China, the United States of America and Argentina are leading linseed-producing countries having cultivars characterised by shorter and much branched stem bearing large seeded pods, whereas, in the temperate regions of China, the Russian Federation and Western Europe, tall plants with much less branching were cultivated as fibre flax [27]. India is the second largest linseed producing country [28] with negligible area under fibre flax cultivation. As a result, this country heavily depends on imports to meet its needs in terms of industrial fibres. Diversified agro-climatic conditions and rich plant genetic resources together with escalating linen import cost have inspired researchers to develop locally adapted fibre flax cultivars in India. The Indian Council of Agricultural Research–Central Research Institute for Jute and Allied Fibres (ICAR-CRIJAF), Barrackpore, Kolkata, India maintains 130 accessions of flax from Asian, European, Australian and American countries. Partial utilisation of these genetic resources paved the way to develop and release the first fibre flax cultivar of India namely “Tiara” during the 11th annual workshop of All India Network Project on Jute and Allied Fibres in 2014 [29]. However, the lack of quantified information about the diversity and the genetic structure of the flax genetic resource seriously limits prospects towards their conservation and enhanced utilization [30].
To this end, initial success in flax diversity assessment by employing both morphological [31–34] and biochemical [35,36] attributes were encouraging. Subsequently, these studies were complemented well by molecular markers like randomly amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), inter simple sequence repeat (ISSR), simple sequence repeat (SSR) and inter-retrotransposon amplified polymorphism (IRAP) [18,37–39]. Presently, marker-based population structure analysis employs methods like FST [40], analysis of molecular variance [41], cluster analysis, principal component analysis and Bayesian analysis [42] etc. across kingdoms. Although some of these methods have powerful algorithms, they all require a suitable marker system reflecting neutral genetic variation to estimate genetic relationships. Microsatellite markers like genomic simple sequence repeats (gSSRs) and expressed sequence tag simple sequence repeats (EST–SSRs) with their ability to enhance the genome wide resolution of genetic variation [43] are one of the most powerful marker systems for population structure analysis of most plant genetic resources [44]. Large numbers of gSSRs [45–49] and EST–SSRs [49–51] have been developed in flax, which can be effectively employed to understand evolutionary and domestication dynamics across morphotypes in the crop.
The literature holds very little information about the population structure of flax and publications are mainly based on the analysis of a few morphological and/or molecular markers across accessions with restricted geographical origin. Therefore, additional research in this line by employing both morphological as well as molecular markers in combination using accessions of diverse origin and morphotypes is highly warranted for a more comprehensive understanding of the evolutionary and domestication processes in the crop. In the present study, 11 morphological traits along with 22 microsatellite-based markers (gSSRs, EST–SSRs) were employed to understand the population structure of 130 flax accessions with diverse growth habits and origins.
2 Materials and methods
2.1 Plant material and layout
A total of 130 accessions (exotic: 75 and indigenous: 55) with diverse growth habits were obtained from the National Active Germplasm Site for Jute and Allied Fibres, ICAR-CRIJAF, Barrackpore, Kolkata, India. These accessions were included in the study after two successive generations of self-pollination by following singleseed descent method to ensure homogeneity of seed lots. The details of these accessions are presented in Table 1. From the homogeneous seed lot of each accession, a part was kept for molecular evaluation and the rest were sown in field for morphological evaluation over two growth seasons. Sowing was done over two seasons (2013–2015) in the experimental fields of ICAR-CRIJAF, Barrackpore, Kolkata, India (22.45°N, 88.26°E; 3.14 above msl) following the standard package of practices to avoid abiotic and biotic stresses that may affect plant growth. During field experiment, each accession was represented by two rows of 3 m, each row at a distance of 30 cm. The plant-to-plant spacing within row was maintained at 5–7 cm by thinning out plants at 35 days after sowing (DAS). All the accessions were sown during the last week of November and harvested at 100 DAS and 125 DAS for fibre and seed quality attributes, respectively.
Details of flax accessions used in present study.
| Fibre type | Intermediate type | Linseed type | |||||||||||||||
| Accession | Source | SPC | Accession | Source | SPC | Accession | Source | SPC | Accession | Source | SPC | Accession | Source | SPC | Accession | Source | SPC |
| LEX001 | Sweden | FTE | LEX003 | Australia | ITE | LEX066 | Australia | ITE | LIN043 | India | ITI | LEX035 | Australia | LTE | LIN028 | India | LTI |
| LEX002 | Sweden | FTE | LEX007 | Canada | ITE | LEX067 | USSR | ITE | LIN044 | India | ITI | LEX036 | Australia | LTE | LIN029 | India | LTI |
| LEX004 | Australia | FTE | LEX008 | USSR | ITE | LEX068 | USSR | ITE | LIN045 | India | ITI | LEX037 | Canada | LTE | LIN052 | India | LTI |
| LEX005 | Australia | FTE | LEX009 | USSR | ITE | LEX070 | Canada | ITE | LIN046 | India | ITI | LEX038 | Canada | LTE | |||
| LEX006 | Australia | FTE | LEX010 | USSR | ITE | LEX071 | Canada | ITE | LIN047 | India | ITI | LEX039 | Canada | LTE | |||
| LEX011 | USSR | FTE | LEX012 | USSR | ITE | LEX072 | Canada | ITE | LIN048 | India | ITI | LEX040 | USSR | LTE | |||
| LEX013 | USSR | FTE | LEX014 | USSR | ITE | LEX073 | Canada | ITE | LIN049 | India | ITI | LEX041 | USSR | LTE | |||
| LEX015 | USSR | FTE | LEX017 | USSR | ITE | LIN001 | India | ITI | LIN050 | India | ITI | LEX042 | USSR | LTE | |||
| LEX016 | Canada | FTE | LEX018 | USSR | ITE | LIN002 | India | ITI | LIN051 | India | ITI | LEX043 | Canada | LTE | |||
| LEX023 | USSR | FTE | LEX019 | USSR | ITE | LIN003 | India | ITI | LIN053 | India | ITI | LEX045 | Canada | LTE | |||
| LEX032 | Belgium | FTE | LEX020 | USSR | ITE | LIN004 | India | ITI | LIN054 | India | ITI | LEX046 | Canada | LTE | |||
| LEX033 | Belgium | FTE | LEX021 | USSR | ITE | LIN005 | India | ITI | LEX048 | Canada | LTE | ||||||
| LEX034 | Holland | FTE | LEX022 | USSR | ITE | LIN007 | India | ITI | LEX049 | Australia | LTE | ||||||
| LEX062 | USSR | FTE | LEX024 | USSR | ITE | LIN008 | India | ITI | LEX050 | Belgium | LTE | ||||||
| LEX063 | USSR | FTE | LEX025 | USSR | ITE | LIN011 | India | ITI | LEX054 | Belgium | LTE | ||||||
| LEX075 | USSR | FTE | LEX026 | USSR | ITE | LIN012 | India | ITI | LEX056 | Argentina | LTE | ||||||
| LEX076 | USSR | FTE | LEX027 | USSR | ITE | LIN014 | India | ITI | LEX057 | Sweden | LTE | ||||||
| LEX064 | USSR | FTE | LEX028 | USSR | ITE | LIN015 | India | ITI | LEX058 | Argentina | LTE | ||||||
| LIM001 | India | FTI | LEX029 | Argentina | ITE | LIN016 | India | ITI | LEX059 | Belgium | LTE | ||||||
| LIM002 | India | FTI | LEX030 | Argentina | ITE | LIN017 | India | ITI | LEX074 | Argentina | LTE | ||||||
| LIM003 | India | FTI | LEX044 | Australia | ITE | LIN018 | India | ITI | LIN009 | India | LTI | ||||||
| LIM004 | India | FTI | LEX047 | Australia | ITE | LIN023 | India | ITI | LIN010 | India | LTI | ||||||
| LIN030 | India | FTI | LEX051 | Belgium | ITE | LIN024 | India | ITI | LIN019 | India | LTI | ||||||
| LIN031 | India | FTI | LEX052 | Belgium | ITE | LIN037 | India | ITI | LIN020 | India | LTI | ||||||
| LIN032 | India | FTI | LEX053 | Belgium | ITE | LIN038 | India | ITI | LIN021 | India | LTI | ||||||
| LIN033 | India | FTI | LEX055 | Argentina | ITE | LIN039 | India | ITI | LIN022 | India | LTI | ||||||
| LIN034 | India | FTI | LEX060 | Belgium | ITE | LIN040 | India | ITI | LIN025 | India | LTI | ||||||
| LIN035 | India | FTI | LEX061 | Belgium | ITE | LIN041 | India | ITI | LIN026 | India | LTI | ||||||
| LIN036 | India | FTI | LEX065 | USSR | ITE | LIN042 | India | ITI | LIN027 | India | LTI |
2.2 Morphological evaluation
For morphological evaluation, data were collected following the descriptor of the International Union for the Protection of New Varieties of Plants (UPOV), Geneva on five randomly selected plants in each accession for 11 traits namely: stem colour (green, red, dark red, green with basal red), flower opening (open, partial open), petal arrangement (free, intermediate, overlapping), flower size (small, medium, large), flower petal colour (white, blue, blue violet), petal shape (circular, circular to pentagonal, pentagonal), colour of distal part of filament (white, blue), anther colour (grey, blue, yellow), boll size (small, medium, large), pod cilia (absent, present), seed coat colour (yellow, brown, deep brown). In the case of qualitative traits, all relevant states of expression were recorded. For recording expression of quasi-quantitative traits an abbreviated scale was used by following UPOV, Geneva descriptors.
2.3 DNA extraction
Genomic DNA was isolated from cotyledonary leaves of 7 days old seedlings using a modified CTAB method [52]. The quality and quantity of extracted DNA were determined using agarose gel electrophoresis (0.8%) and a spectrophotometer (Eppendorf), respectively.
2.4 gSSR and EST-SSR analysis
A total of 21 gSSRs [47,49] and 10 EST-SSRs [50] loci linked to the cell wall biochemical synthesis pathway were selected for the present study. Finally, 15 gSSRs and 7 EST-SSR markers exhibiting reliable amplification were used for further analysis (Table 2). PCR reactions were performed in 25 μl reaction mixture containing 0.2 mM of dNTPs, 0.5 μM each of forward and reverse primers, 0.8 unit of Taq DNA polymerase (Invitrogen) and 0.5 μl MgCl2 (2.5 mM) in 1X PCR buffer (10 mM Tris–HCl buffer, pH 8.0, 0.5 mM KCl). PCR amplification was carried out with a hot start for 5 min at 95 °C followed by 35 cycles at 95 °C for 30 s, 1 min at the annealing temperature and 72 °C for 1 min with a final extension at 72 °C for 10 min in SimpliAmp™ Thermal Cycler (Applied Biosystems, USA). The PCR amplification products were stained with ethidium bromide and ran in 2.0% agarose gel in 1X TBE buffer (1.5–2.0 h at 3.82 V/cm). The size of the amplicon was determined using a 100 bp DNA ladder (product ranged from 100 to 3000 bp) (Genaxy, India) and visualized as well as photographed under a gel documentation unit (Bio-Rad, USA).
2.5 Data analysis
The expression of morphological traits (a total of 28 alleles) was binary-coded as 1 for presence or 0 for absence in each accession [53,54]. Owing to the self-pollinating reproductive behaviour of flax, any heterozygosity was not expected for these traits. For molecular marker (gSSR and EST-SSR) profiles, each band was scored as for the presence (1) or for the absence (0). Only the distinct, reproducible and well resolved bands were scored. Before analyzing the data, the flax accessions were classified into three broad groups (based on morphotypes and technical height) and each group into two sub-groups (based on origin): fibre type (flax; convar. elongatum) (29 accessions: 18 exotic; 11 indigenous), intermediate type (convar. usitatissimum) (69 accessions: 37 exotic; 32 indigenous) and oilseed type (linseed; convar. mediterraneum) (32 accessions:20 exotic; 12 indigenous) by adopting Kulpa and Danert [55]. Thus, a total of six sub-populations (fibre type exotic, fibre type indigenous, intermediate type exotic, intermediate type indigenous, linseed type exotic and linseed type indigenous) were constituted a priori and subsequent analyses were performed considering this classification. Further, genetic diversity parameters namely; Nei's unbiased genetic identity (I) and genetic distance (D), expected heterozygosity (He), Shannon information index (SI), number of effective alleles (Ne), and percentage of polymorphism (P) [56,57] were determined for each sub-population using GenAlEx v. 6.1 software [58]. The software helped to partition the molecular variance among and within sub-populations and test their statistical significance through the analysis of molecular (both morphological and molecular) variance (AMOVA) [41] using 1000 permutations. Furthermore, GenAlEx v. 6.1 software was also used for principal coordinate analysis (PCoA) based on different makers. Assuming an Hardy–Weinberg equilibrium, POPGENE version 1.31 [59] software was used to estimate total genetic diversity (Ht), intrasub-population genetic diversity (Hs). Intersub-population genetic diversity (Dst), genetic differentiation coefficient (Gst), were calculated according to de Vicente et al. [60] as follows; Dst = Ht–Hs and Gst = Dst/Ht.
Population structure analysis based on different markers (morphological traits, gSSRs, EST–SSRs and combined) was done by a Bayesian model-based clustering algorithm of STRUCTURE 2.3 [42,61] software. The STRUCTURE 2.3 software was run using putative population origin with parameters as follows: burn-in periods = 5000, MCMC replicates = 50,000, admixture ancestry model, allele frequency correlated (K = 1 to 10, iterations = 10). To determine the optimum value of K, the results of STRUCTURE 2.3 software were used as input in Structure Harvester online software [62]. Based on Evanno et al.’s delta K method [63], the optimum number of clusters was identified for different markers. After that, the STRUCTURE 2.3 software was run at desired K values for 100,000 burn-in periods and 100,000 MCMC replicates. Thereafter, a structure diagram was prepared in the same software.
3 Results
3.1 Genetic diversity of population
Different diversity indices were estimated for three populations (fibre type, intermediate type and linseed type) and six sub-populations of flax using morphological traits (11), gSSR (15) and EST-SSR (7) markers that produced 28, 47 and 13 loci, respectively (Table 3). At the species level, the highest diversity was exhibited by gSSR markers (SI = 0.46; He = 0.31; P = 85.11%). However, morphological markers recorded the lowest number of alleles (1.6), but higher polymorphic loci (76.79%) than EST-SSR (73.08%).
Diversity parameters estimated based on molecular (EST-SSR, gSSR) and morphological traits in Flax.
| Population | EST-SSR | gSSR | Morphological | Combined | ||||||||||||||||
| Na | Ne | SI | He | P (%) | Na | Ne | SI | He | P (%) | Na | Ne | SI | He | P (%) | Na | Ne | SI | He | P (%) | |
| Population level | ||||||||||||||||||||
| Fibre type | 1.85 | 1.38 | 0.38 | 0.24 | 84.62 | 1.89 | 1.52 | 0.45 | 0.30 | 89.36 | 1.93 | 1.47 | 0.43 | 0.28 | 92.86 | 1.90 | 1.48 | 0.43 | 0.29 | 89.77 |
| Intermediate type | 2.00 | 1.46 | 0.45 | 0.29 | 100.00 | 2.00 | 1.64 | 0.54 | 0.37 | 100.00 | 1.93 | 1.38 | 0.38 | 0.24 | 96.43 | 1.99 | 1.53 | 0.47 | 0.31 | 98.86 |
| Linseed type | 1.92 | 1.55 | 0.49 | 0.33 | 92.31 | 1.94 | 1.63 | 0.53 | 0.36 | 93.62 | 1.86 | 1.33 | 0.33 | 0.21 | 85.71 | 1.91 | 1.52 | 0.46 | 0.31 | 90.91 |
| Sub-population level | ||||||||||||||||||||
| FTE | 1.23 | 1.08 | 0.11 | 0.06 | 38.46 | 1.68 | 1.42 | 0.38 | 0.25 | 76.60 | 1.86 | 1.48 | 0.44 | 0.29 | 92.86 | 1.76 | 1.39 | 0.36 | 0.23 | 76.14 |
| FTI | 1.62 | 1.61 | 0.47 | 0.33 | 76.92 | 1.81 | 1.49 | 0.43 | 0.29 | 80.85 | 1.43 | 1.43 | 0.37 | 0.25 | 67.86 | 1.76 | 1.49 | 0.41 | 0.28 | 76.14 |
| ITE | 1.77 | 1.42 | 0.40 | 0.26 | 84.62 | 1.96 | 1.62 | 0.53 | 0.36 | 97.87 | 1.75 | 1.40 | 0.37 | 0.24 | 85.71 | 1.92 | 1.52 | 0.46 | 0.30 | 92.05 |
| ITI | 1.84 | 1.40 | 0.39 | 0.25 | 84.62 | 1.89 | 1.62 | 0.52 | 0.35 | 93.62 | 1.93 | 1.34 | 0.35 | 0.22 | 96.43 | 1.92 | 1.50 | 0.45 | 0.29 | 93.18 |
| LTE | 1.77 | 1.49 | 0.42 | 0.28 | 76.92 | 1.85 | 1.60 | 0.49 | 0.34 | 87.23 | 1.39 | 1.29 | 0.28 | 0.18 | 64.29 | 1.78 | 1.48 | 0.42 | 0.28 | 78.41 |
| LTI | 1.69 | 1.50 | 0.42 | 0.29 | 76.92 | 1.72 | 1.48 | 0.41 | 0.27 | 74.47 | 1.25 | 1.28 | 0.25 | 0.16 | 53.57 | 1.68 | 1.42 | 0.36 | 0.24 | 68.18 |
| Species level | 1.65 | 1.42 | 0.37 | 0.25 | 73.08 | 1.82 | 1.54 | 0.46 | 0.31 | 85.11 | 1.6 | 1.37 | 0.34 | 0.22 | 76.79 | 2.00 | 1.52 | 0.47 | 0.31 | 100.00 |
At the morphotype level, intermediate type flax showed the highest value for all diversity parameters by combining morphological traits with gSSR and EST-SSR markers (Table 3). gSSR markers and morphological traits revealing maximum and minimum diversity across parameters, respectively. However, maximum Shannon's information index (SI) and expected heterozygosity (He) expressed by gSSR in all the three populations namely fibre type(SI = 0.45, He = 0.30), intermediate type (SI = 0.54, He = 0.37) and linseed type (SI = 0.53, He = 0.36). Among all sub-populations across all markers the highest number of alleles, Shannon's information index and polymorphic loci were recorded by gSSR in intermediate exotic population (Na = 1.96, SI = 0.53 and P = 97.87%, respectively). The lowest value for these parameters was recorded for fibre type exotic accessions for EST-SSR markers (Na = 1.23, SI = 0.11 and P = 38.46%, respectively). The maximum number of alleles in fibre type exotic (Na = 1.86) and intermediate type indigenous (Na = 1.93) sub-population were recorded by morphological traits. In fibre type indigenous, intermediate type exotic, linseed type exotic and indigenous sub-populations gSSR markers recorded the highest number of observed alleles (Na = 1.81, 1.96, 1.85 and 1.72, respectively). The percentage polymorphic loci across sub-populations followed the same trend, except for linseed-type indigenous accessions that recorded highest value for EST-SSR markers (P = 76.92%). By taking all markers into account, intermediate exotic population represent the sub-population with maximum number of total alleles (Na = 1.92), effective alleles (Ne = 1.52) and Shannons’ information index (SI = 0.46) with considerably high expected heterozygosity (He = 0.30).
Primer sequence of gSSR and EST-SSR markers used in the study.
| Marker | gSSR primers (5′-3′) | Marker | EST-SSR primers (5’-3’) |
| LU2 | F- TCCGGACCCTTTCAATATCA | COMT | F- TATCCAAGCACCCTTCCATT |
| R- AACTACCGCCGGTGATGA | R- GTTCCTCGTCGCCAGGCTCGTGT | ||
| LU3 | F- GCTCGTGATCTCCTTCATCC | CCoAOMT1 | F- TCGGCGTCTACACGGGTTACTCC |
| R- AAAACCACGTCCAGATGCTC | R- TGTTGTCGTACCCGATCACTC | ||
| LU4 | F- TTATTTCCGGACCCTTTCAA | 4CL2 | F- GGGATACGGAATGACAGAGG |
| R- AAACTACCGCCGGTGATGAT | R- CGGTGTGTAACCACCCTTGTTTGTC | ||
| LU5 | F- GTCACTGGGTGTGTGTTTGC | ACTIN | F- CGACTGCTGAAGGGAAATTGT |
| R- AGCAGAAGAAGATGGCGAAA | R- CTGCTGGAAGGTGCTGAGGGAA | ||
| LU6 | F- CCCATTTCTACCATCTCCTT | F5H1 | F- CGTCGGAGAGCTTATTTTCA |
| R- CAACAGCGGAACTGATGAAA | R- AGCTTTGATGTTGTCCTTGG | ||
| LU7 | F- CATCCAACAAAGGGTGGTG | F5H2 | F- CCACCAAGATGAATGACCAG |
| R- GGAACAAAGGTAGCCATGA | R- TGTAGTGGTGGTGGAGTCAGCTGC | ||
| LU9 | F- TTGCGTGATTATCTGCTTCG | F5H3 | F- CGCTGGCCGGGCGTTGAACG |
| R- ATGGCAGGTTCTGCTGTTTC | R- ATGTCGGAGATAGGATCGTC | ||
| LU10 | F- GCCTAAAGCTGATGCGTTTC | ||
| R- TGTCAGGCTCCTTCTTTTGC | |||
| LU11 | F- ATGGCAGGTTCTGCTGTTTC | ||
| R- TTGCGTGATTATCTGCTTCG | |||
| LU16 | F- TTATTCTTGCCTGCCAATCG | ||
| R- TCCAGCTCTTGCTCGTTCTT | |||
| LU17 | F-GTTTGGTGTTTGGGTGCTTT | ||
| R-TTCGGTTGTAGCATCCATCC | |||
| LU18 | F- AGAGGCGGAGGGCATTAC | ||
| R- TTGGAGAGTTGGAATCGAGA | |||
| LU27 | F- GTTTGAGAAGAGGGCATCCA | ||
| R- GTTGGGGTGAAGAGGAACAA | |||
| LU32 | F- ACGCGTAAACTTTCCGTTTC | ||
| R- ATAATGTCGGCTGCTTCTGC | |||
| LU35 | F- CCAACGGATCATCCTCTAGC | ||
| R- GGAACAGAAAGGGGAAAGGAA |
AMOVA analysis based on morphological, EST-SSR and gSSR markers underlined significant difference among fibre type, intermediate type and linseed type (Table 4). In comparison to morphological and gSSR (фPT = 0.09 and 0.06, respectively at p = 0.01) EST-SSR markers (фPT = 0.12, p = 0.01) better differentiate flax populations. All the three marker system revealed higher intrasub-population variations than intersub-population variations. Nei's unbiased measures of genetic distance (D) and genetic identity (I) of six sub-populations for morphological, EST-SSR and gSSR markers were calculated (Table 5). The analysis indicates a maximum genetic distance (D = 0.11) between fibre-type exotic sub-populations and linseed-type exotic sub-populations compared to other sub-populations based on EST–SSR markers. The lowest genetic distance was recorded between intermediate exotic type and indigenous sub-populations for gSSR (D = 0.02) and morphological markers (D = 0.006). However, for EST-SSR markers, linseed-type indigenous and intermediate-type exotic sub-populations revealed almost no genetic distance (Fig. 1).
Analysis of molecular variance (AMOVA) within/among sub-populations of flax.
| Marker | Source | df | Sum of square | Estimated variation | Percentage of total variation | ϕPT a | P-value |
| Morphological | Among sub-population | 5 | 53.578 | 0.350 | 9 | 0.09 | 0.01 |
| Within sub-population | 124 | 426.145 | 3.437 | 91 | |||
| EST-SSR | Among sub-population | 5 | 26.047 | 0.186 | 12 | 0.12 | 0.01 |
| Within sub-population | 124 | 166.453 | 1.342 | 88 | |||
| gSSR | Among sub-population | 5 | 80.069 | 0.422 | 6 | 0.06 | 0.01 |
| Within sub-population | 124 | 896.700 | 7.231 | 94 |
a ϕPT, population differentiation: proportion of the variance among the sub-populations relative to the total variance. The probability of obtaining an equal or lower фPT value was determined by 999 random permutations.
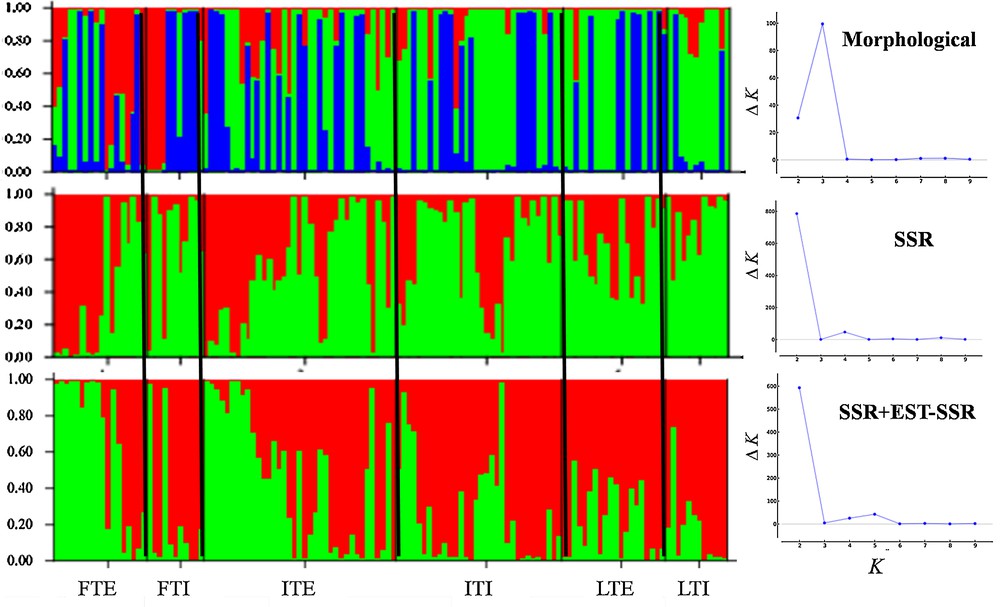
Population structure flax germplasm accessions based on different markers. Please refer to Table 1 for abbreviation details.
3.2 Population structure
For structure morphological, gSSR, EST-SSR+gSSR markers were used (Fig. 1). The ΔK values for the optimum number of clusters in the flax population were determined at K = 3 and 2 for morphological markers and gSSR/EST-SSR+gSSR markers, respectively by following the method of Evanno et al. [63]. Within cluster accessions were represented by unique colour while accessions with two different colours indicating admixed forms. The results of the EST-SSR marker-based independent analysis are not presented here, due to the limited number of loci. Morphological markers grouped the accessions into three clusters with ΔK = 3. The second cluster with 66 accessions was the largest and mainly comprises intermediate-type accessions from Indian and exotic origin (25.8% each). Cluster-I (17 accessions) are mainly constituted by fibre-type exotic accessions from West European countries. The remaining accessions grouped into cluster-II (47 accessions). gSSR marker-based clustering grouped fibre-type exotic (20%) and intermediate type (Indian = 20.0%, exotic = 38.2%) together in cluster-I. Most of the linseed type exotic (17.3%) and intermediate type (Indian = 28.0%, exotic = 21.3%) belonged to cluster-II. Further, EST-SSR+gSSR markers grouped accessions into two clusters. Cluster-II was dominated by exotic accessions of intermediate and fibre type (∼50% and ∼25%, respectively), while cluster-I represented comparatively a mixed group with intermediate (Indian∼31%, exotic∼23%) and linseed-type exotic (∼21%) flax.
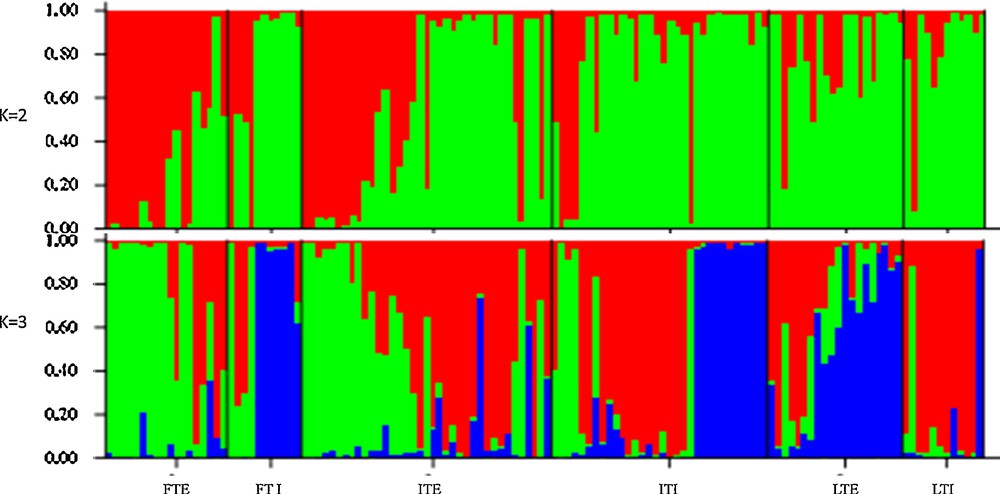
Population structure of flax germplasm accessions based on combined markers. Please refer to Table 1 for abbreviation details.
The combined analysis of morphological, EST-SSR and gSSR markers grouped exotic accessions into 2 and 3 clusters at K = 2 and K = 3, respectively (Fig. 2). Initially, with two clusters populations (K = 2), exotic accessions of fibre and intermediate type (88.9% and 64%, respectively) were distinguished from the rest of the flax genotypes and grouped into a separate cluster. When one more subgroup was allowed (K = 3), Indian accessions of fibres were (63.4%) distinguished and constituted a separate cluster with the Indian intermediate type (48.7%). Cluster-II was predominantly constituted by Indian accessions of linseed type (82.16%). However, Indian intermediate type and exotic linseed type accessions were distributed among the above two clusters.
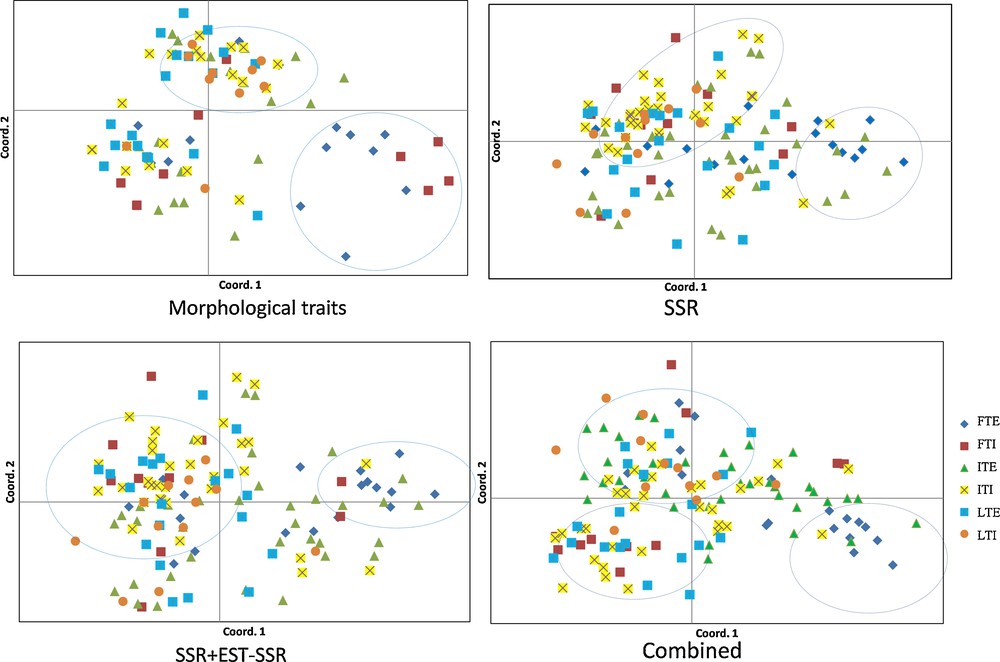
Principal coordinate analysis (PCoA) plot of flax germplasm populations. Please refer to Table 1 for the abbreviation details.
The principal coordinate analysis (PCoA) plots for morphological, gSSR, EST-SSR+gSSR and combined (morphological + EST-SSR + gSSR) markers well complement findings of the structure analysis (Fig. 3). In all cases, exotic fibre type and Indian linseed type grouped distinctly. Morphological and EST-SSR+gSSR distinguished exotic fibre type from Indian linseed type. gSSR based analysis genetically distinguish Indian intermediate type, Indian linseed type and exotic fibre-type flax accessions. However, the combined marker based PCoA plot clearly depicts three clusters. The first axis differentiated fibre-type from linseed-type accessions, except very few outliers, while the second axis separated Indian accessions of fibre type from exotic one. Exotic fibre type accessions constituted a cluster together with the exotic intermediate type. In contrast, Indian fibre type accessions form a separate cluster with Indian intermediate type accessions. Part of the exotic and Indian accessions of intermediate type exhibited an overlap indicating continuity between fibre and linseed type.
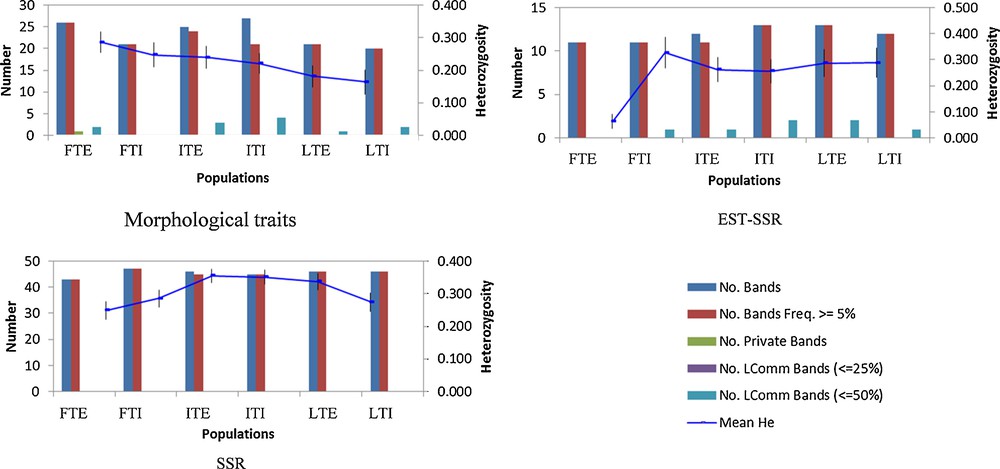
Heterozygosity in different populations of flax. Please refer to Table 1 for abbreviation details.
4 Discussion
In the present study, morphological traits were employed to complement microsatellite markers for deciphering genetic diversity and population structure as they were reported as being equally effective for the cause [53,54]. The result highlighted the efficiency of these morphological traits in distinguishing morphotypes namely; fibre type, intermediate and linseed type in flax. In spite of the successful use of morphological traits for morphotypes discrimination, consistent lower values of polymorphic loci percentage (76.79%) were observed in comparison to the employed gSSR primers (85.11%). However, they performed almost at par with EST-SSR primers (73.08%). Hence, the efficiency of a given trait/primer depends on the number of states/fragments it generates as well as on their frequency. The higher number of loci (47) revealed by gSSR markers in comparison to the number of states/loci found for the morphological traits and EST-SSR markers (28 and 13, respectively) explains the observed differential polymorphic loci percentage of the three markers in the study. Similar estimates of genetic relationships among flax sub-populations were obtained for morphological traits, gSSR and EST-SSR markers in the present study. The limitation of morphological traits and EST-SSR markers in revealing genetic diversity and genealogy perhaps stems from the fact that they are functional markers exploring the genetic diversity of a population for genes that may be of agronomic importance [65].
Thus the study suggests that, in spite of the powerfulness of microsatellite markers in depicting genetic relationships, they should not be seen as a substitute of morphological traits [66] as each marker system measures different aspects of genetic variability [67]. In the present study, morphological traits reveal the presence of certain private (rare) alleles that are neither observed in gSSR nor in EST-SSR markers (Fig. 4). Other findings highlighted implications of domestication on the loss of rare alleles in a number of crops, including soybean [68]. Therefore, exotic fibre type accessions with such rare alleles are an asset for Indian flax breeding programmes. Over all, in the light of present results and earlier evidences, a combination of marker systems is recommended to be employed in a complementary manner in order to provide relatively complete information of the genetic relationships in plants.
Morphological markers primarily differentiate fibre-type exotic accessions from the rest of the accessions, while gSSR could differentiate fibre exotic and linseed-type exotic accessions. However, EST-SSR and gSSR markers put together most of the exotic fibre and intermediate type. The relative efficiency of microsatellite markers over morphological markers may be attributed to the neutral nature of DNA markers [69–71]. To further enhance the resolution of population structure combined analysis using morphological, EST-SSR and gSSR markers, particularly at K = 3, grouped flax accessions in agreement with their broad morphotype classification and geographical origin. The result highlights the strength of functional markers like morphological traits and EST-SSR markers in augmenting neutral markers (gSSR) during phylogenetic studies of crop-like flax having complicated domestication history [64]. These reports are of special significance for morphological traits given their cost effectiveness and time efficiency while handling large populations. Already their effectiveness in genealogy analysis and genetic diversity assessment are being recognized across crops [53,54].
The PCoA analysis was performed by using genotypic data of morphological traits, gSSR, EST-SSR+gSSR and morphological+gSSR+EST-SSR. Finding that all the markers employed in isolation or in combination successfully differentiated exotic fibre-type flax accessions supports the view that all modern exotic fibre type originated from Eastern Europe and may have a common ancestor [21,24]. However, grouping of Indian linseed type in a separate cluster may be due to small sample size as they represent a few accessions maintained together with fibre type accessions at ICAR-CRIJAF, Barrackpore, Kolkata, India. Hence, they reflect only a limited spectrum of Indian linseed genetic diversity. Finally, based on combined (morphological+gSSR+EST-SSR) markers entire flax sub-populations can be broadly groped into two group namely fibre type and linseed type. This is in conformity with findings of Allaby et al. [21]. Further, the linseed type with presence in more than one cluster appears genetically more diverse than the fibre type (Fig. 3). This is perhaps the outcome of restricted geographical distribution of fibre flax in certain pockets, particularly since the 17th century when cotton replaced flax as the prime fibre crop [1]. In contrast, the popularity of flax for seed oil continued to increase over years, particularly after the industrial revolution [21] that led to widespread cultivation and distribution across continents. The Indian fibre flax appears genetically distinct from exotic fibre type accessions and might have originated from independent domestication events or years of reproductive isolation coupled with evolutionary forces like mutations, selection, recombination, genetic drift and human interference. A similar phenomenon was recorded in flax accessions from West and South East Asia [72]. The finding need to be supplemented with further studies involving more fibre flax accessions from diverse origins, particularly from Asian countries, so as to draw precise conclusions.
5 Future perspective
The genus Linum requires end use specific breeding programme to meet diversified applications. To this aim, the information derived from the present study will prove helpful for developing precise selection indices for the identification of superior lines in the flax improvement programme. The identified groups based on both morphological and molecular markers can be an ideal base material to develop a broad spectrum breeding populations suitable for diversified end uses.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgement
The authors are thankful to the director, ICAR–CRIJAF, Barrackpore, Kolkata, India for providing research facilities. Valuable contributions of Dr. D. Gupta and Dr. S.K. Hazra (Ex-Principal Scientist, ICAR-CRIJAF) in collection/exchange and maintenance of flax germplasm accessions from diverse sources are deeply acknowledged. Field technical assistance provided by Mr. O.P. Choudhary and Dileep Kumar is duly acknowledged. Authors are thankful to anonymous reviewers who's valuable suggestions helped us to improve the manuscript.


