1 Introduction
Metals and metalloids are one of the major environmental pollutants. The presence of high concentrations of these elements in the environment can cause several harmful and detrimental effects to human health and poses a serious threat to the environment [1]. These metals are either present in bioavailable or non-bioavailable forms. Toxicity of metals and metalloids is due to their interaction with proteins resulting in the inhibition of metabolic processes [2,3]. These toxic metals and metalloids find their way to the food chain through industrial effluents that contaminate water and soil. According to EPA, arsenic [As, both As(V) and As(III)], chromium (Cr), cadmium (Cd), copper [Cu], lead (Pb), mercury (Hg), nickel (Ni) and zinc (Zn) are most hazardous contaminants [4]. As can cause multiple disorders, including cancers of various organs, and it plays roles in multifactorial diseases such as cardiovascular problems. Bioremediation is an environment-friendly alternative of chemical and physical remediation technologies. Microorganisms and their products have proved more effective than physical and chemical methods of remediation [5]. Microbes exposed to toxic metals develop resistance against toxicants [6]. Such microorganisms evolve various strategies to survive in stressful conditions. Microorganisms can transform toxic forms of metals into less or non-toxic forms. They may also use metals for their structural, catalytic functions and as electron acceptors, though at low concentrations [7]. Efflux of metal ion outside of the cell, accumulation and complexation of metal ion inside the cell, oxidation/reduction of the toxic form of metal to less toxic form and modification of the cell components to lessen metal sensitivity are some of the mechanisms employed by microbial cells to cope with metal toxicity [8,9]. Bacteria present in the rhizosphere have also been reported to resist toxic metals and metalloids such as As [10].
The purpose of the present study was to investigate the multi-metal resistance ability of bacteria. Sampling was done in Kasur city, which has been a major hub of tanneries for decades. As the major pollutants of these industries are As and Cr, therefore the abilities of bacterial isolates to oxidize/reduce As and Cr was also studied. As many metal resistance genes are plasmid borne, therefore, it was also analysed whether this multi-metal resistance ability is chromosome or plasmid mediated. The multi-metal-resistant bacteria were identified through 16S rRNA gene sequencing.
2 Material and methods
2.1 Sampling and isolation of multiple metal-resistant bacteria
Stock solutions of As(V) (Na3AsO4), As(III) (Na2AsO2), Cr(VI) (K2CrO4), Cu (Cu2SO4), Co (CoCl2.6H2O), Cd (CdCl2), Hg (HgCl2), Se (Na2SeO3), Pb (PbCl2), Ni (NiCl2), and Zn (ZnSO4) were prepared and filter sterilized. Contaminated soil samples were collected from the wastewater treatment plant at Kasur, Pakistan (31°05′40.0″N, 74°28′04.7″E), in sterile containers. The wastewater treatment plant receives wastewater from local industries, which are mostly tanneries. The soil samples were transported to the laboratory; pH and temperature were recorded. Multi-metal-resistant bacteria were isolated using an enrichment culture technique by adding 1.0 g of soil in a tryptic soya broth (TSB) containing 1.0 mM each of As(III), As(V), Cr, and Se. After 24 h of incubation at 37 °C and 150 rpm, 1.0 mL of the culture broth was transferred into a fresh TSB flask containing the above-mentioned metals as well as 1.0 mM of Zn, and was incubated at 37 °C under 150-rpm agitation. The above process was repeated until the culture was supplemented with As(III), As(V), Cd, Co, Cu, Pb, Se, Ni, Zn (1.0 mM each) and Cr and Hg (0.5 mM each) [11]. Cultures from every enrichment flask were serially diluted and plated on Luria–Bertani (LB)-agar plates. CFU·g−1 was counted and diverse colonies from the last enrichment culture (containing all the metals) were purified by repeated quadrant streaking on LB-agar.
2.2 Determination of metal resistance and minimum inhibitory concentration
For the determination of the metal resistance of isolates, mineral agar media plates [12] were prepared containing As(III), As(V), Co, Cd, Cu, Se, Ni, Pb or Zn ranging from 1.0 to 30 mM. For Cr and Hg, concentrations ranging from 0.5 to 30 mM were used [11]. Bacterial isolates were cultured on these plates. After 72 h of incubation at 37 °C, the presence or the absence of growth was recorded.
2.3 Optimization of the isolated bacteria
Selected isolates were characterized morphologically and biochemically [13] (see supplementary material). The optimal growth conditions (pH and temperature) of the isolates were also determined. Overnight bacterial cultures were standardized at 0.2 OD600nm and inoculated in the LB broth, followed by incubation at 15 °C, 25 °C, 37 °C and 45 °C (pH 7.0). For optimal pH determination, LB broth in four different batches was prepared and set at pH 3, 5, 7 and 9. The medium was inoculated with the same standardized cultures and incubated at 37 °C. After 24 h of incubation, the optical densities were recorded at 600 nm.
2.4 Determination of antibiotic resistance
The bacterial isolates were cultured on LB-agar media plates supplemented with antibiotics such as ampicillin (1.0 μg·mL−1), chloramphenicol (3.0 μg·mL−1), erythromycin (1.5 μg·mL−1), kanamycin (3.0 μg·mL−1) and tetracycline (3.0 μg·mL−1) and incubated at 37 °C. After incubation, the plates were observed for bacterial growth.
2.5 Estimation of As-oxidation/reduction by bacterial isolates
The extent of As-oxidation/reduction was assessed by the assay reported by Cummings et al. [14], with minor modifications. Overnight bacterial cultures were centrifuged, and pellets were washed with and resuspended in 0.9% NaCl (pH 7.0). The suspension was divided into two sets, one was supplemented with 1000 μM of As(V), while the other one was supplemented with 500 μM As(III) and incubated at room temperature for 5 h. Supernatants were collected by centrifugation before the start of incubation as well as after incubation.
To quantify As(V) reduction into As(III) or As(III) oxidation into As(V), 100 μL of each supernatant was acidified with 100 μL of 24 mM HCl. One hundred microliters of the acidified samples were added to 900 μL of the colouring reagent, which contained 9.9 g of ascorbic acid, 0.132 g of potassium antimony tartrate, 6 g of ammonium molybdate, and 500 mL of 5 N sulphuric acid per litre. The reaction mixture was heated at 78 °C for 20 min on water and incubated on ice for 5 min. The absorbance was determined at 865 nm using a spectrophotometer (Cecil CE 7200).
As this assay measures As(V) rather than As(III), the samples were oxidized to confirm As(V)-reduction. As(III) produced in the samples [due to As(V) reduction] was oxidized back to As(V) by addition of 100 μL of 5 mM KIO3 and 48 mM of HCl. The mixture was heated at 78 °C for 5 min and 100 μL of the oxidized samples were added to 900 μL of colouring reagent and processed as described above. As(V) reduction was estimated by subtracting the As(V) concentration of oxidized samples from the As(V) concentration of unoxidized samples. The concentration of reduced As(V) or oxidized As(III) was calculated by generating a standard curve.
2.6 Genomic DNA isolation and 16S rRNA sequencing
Genomic DNA was isolated using the method described by Wilson [15]. The 16S rRNA gene was amplified using the universal primers 8F (AGAGTTTGATCCTTGGCTCAG) and 1492R (GCYTACCTTGTTACGACTT). Amplicons were confirmed using agarose gel electrophoresis and were sequenced from Macrogen (Korea). The quality of the base calling was checked in FinchTV (Geospiza) and contigs were created through BLAST tool of NCBI. Closest matching sequences from the BLAST results were downloaded, aligned, and a neighbour-joining tree was made in MEGA 5 [16].
2.7 Plasmid isolation and transformation
Plasmid isolation was carried out using the GF-1 plasmid DNA extraction kit (Vivantis, USA) following the manufacturer's instructions, and the yield was confirmed using agarose gel electrophoresis. Plasmids isolated from multi-metal-resistant bacteria were used to transform Escherichia coli DH5α. CaCl2-based method was used to make competent cells of E. coli DH5α [17]. DH5α was cultured in a LB broth to 0.6 OD600 nm and cells harvested by centrifugation at 4 °C. The pellets were suspended in 0.1 M MgCl2 and incubated on ice for 10 min. The suspension was centrifuged at 4 °C and pellets were resuspended in 0.05 M CaCl2 followed by incubation on ice for 20 min. After incubation, the pellets were obtained by centrifuging the suspension at 4 °C. The pellets were dissolved in 0.5 M CaCl2 with glycerol solution. Competent cells (200 μL) were mixed with plasmid (10 μL), and the mixture was incubated for 30 min on ice. The reaction mixture was given a heat shock at 42 °C using a water bath for 45 s and incubated again on ice for 2.0 min. The reaction mixture was provided with 1.0 mL of LB broth and incubated at 37 °C for 30 min. After incubation, transformants were screened on LB-agar plates containing the metals mentioned previously.
2.8 Plant growth promoting abilities
Indole-3-acetic acid (IAA) production, phosphorous solubilisation and hydrogen cyanide (HCN) production by the bacterial isolates was determined. Bacterial cultures MX-2 and MX-6 were inoculated in an L-broth supplemented with l-tryptophan. After 72 h of incubation at 37 °C, 120 rpm, 100 μL of the supernatant were mixed with 200 μL of Salkowski's reagent [18] in a microtitre plate. After 1.0 h of incubation in the dark, the optical density was recorded at 535 nm using a Biotek Epoch plate reader. The concentration of IAA in the culture was measured using standard curve achieved using IAA standards.
The phosphate solubilisation ability of MX-2 and MX-6 was determined by culturing the bacteria in the phosphate agar media of the National Botanical Research Institute (NBRI) [19]. The plates were incubated for 5 to 7 days at 37 °C. The presence of clear zones around bacterial growth indicated phosphate solubilisation.
To evaluate the ability of isolates to produce hydrogen cyanide, bacteria were inoculated on nutrient agar media plates supplemented with glycine (4 g L−1) [20]. After 4 days of incubation at 37 °C, the development of a yellow to orange colour was observed.
2.9 Plant–microbe interaction
The experiment to study the phytostimulatory effect of the bacterial isolates on plant growth in the presence and absence of metals was analysed in pots [5,11]. Seeds of Vigna radiata (mung bean plant) were washed with autoclaved distilled water and were surface-sterilized with 0.1% HgCl2 for 2 min followed by washing thrice with distilled water. Pots were filled with sieved soil and divided into four sets; plants, plants + bacteria, plant + metals and plants + metals + bacterial strains.
For inoculation, fresh cultures of MX-2 and MX-6 were taken and bacterial suspension was prepared in a normal saline solution (0.9% NaCl). Surface-sterilized seeds were soaked in bacterial suspension (0.5 OD600 nm) for 20 to 25 min, and eight seeds were sown at equal distances in each pot. Thinning was done after germination, leaving five plants in each pot. A metal solution was used for watering where needed. Each pot contained only one of the metals (0.05 mM Hg, Cr, 0.1 mM As(III), As(V), Co, Cd, Cu, Se, Ni, Pb or Zn). The pots were placed at a temperature of 25 ± 1 °C during a 12-h photoperiod. After two weeks, root and shoot lengths, number of leaves and roots, wet and dry weights were recorded. There were at least ten pots of each condition, the mean was taken and standard deviation was applied.
2.10 Statistical analysis
All the experiments were performed in triplicates unless mentioned otherwise. The mean of the results was plotted and the standard deviation was shown as error bars. The plant–microbe interaction data were analysed and compared statistically by using ANOVA (analysis of variance) followed by two-tailed t-test post-hoc analysis (P = 0.05). Bonferroni correction was also applied to adjust the threshold of significance. The analysis was done with Microsoft Excel 2013.
3 Results
3.1 Isolation of multi-metal-resistant bacteria
The pH of the sample was 7.0, while the temperature was 40 °C. Bacteria were isolated using an enrichment culture technique and enumerated using a serial dilution technique on LB-agar plates. Six bacterial isolates (MX-1, MX-2, MX-3, MX-4, MX-5 and MX-6) resistant to all the metals tested [1.0 mM As(III), As(V), Cd, Cu, Co, Se, Pb, Ni, Zn and 0.5 mM Cr and Hg] were selected for further analysis.
The CFU count in the presence of As(III), As(V), Cr and Se was found to be 3.2 × 105. The CFU count was found to be 3.7 × 104 when As(III), As(V), Cr, Se and Zn were added to the next enrichment flask. CFU further decreased to 2.8 × 104 when supplemented with As(III), As(V), Cr, Se, Zn and Cd. Further enrichment of bacteria in the presence of As(III), As(V), Cr, Se, Zn, Cd and Ni resulted in a CFU count of 2.5 × 103. With the addition of Co, the CFU count dropped further to 2.2 × 103. Further addition of Pb and Cu resulted in the decrease of CFU count to 2.8 × 102. In the presence of As(III), As(V), Cr, Se, Cu, Zn, Co, Cd, Ni, Pb and Hg, the final CFU count was 1.2 × 102.
3.2 Characterization and optimization of bacteria
Selected bacteria were subjected to morphological and biochemical analyses (see supplementary material). The temperature and pH optima for these isolates (for which the best growth was observed) were 37 °C and 7.0, respectively. The lesser growth was observed at pH 3 and at 45 °C.
3.3 16S rRNA gene analysis
Based on morphological and biochemical characteristics as well as 16S rRNA gene sequencing, the isolate MX-6 was identified as Pseudomonas sp. The neighbour-joining tree of 16S rRNA sequences of MX-6 and closely matched NCBI BLAST results was made in MEGA 5.0 to confirm the identity of the bacteria (Fig. 1). The sequence was submitted to NCBI GenBank with the accession number KU195676.
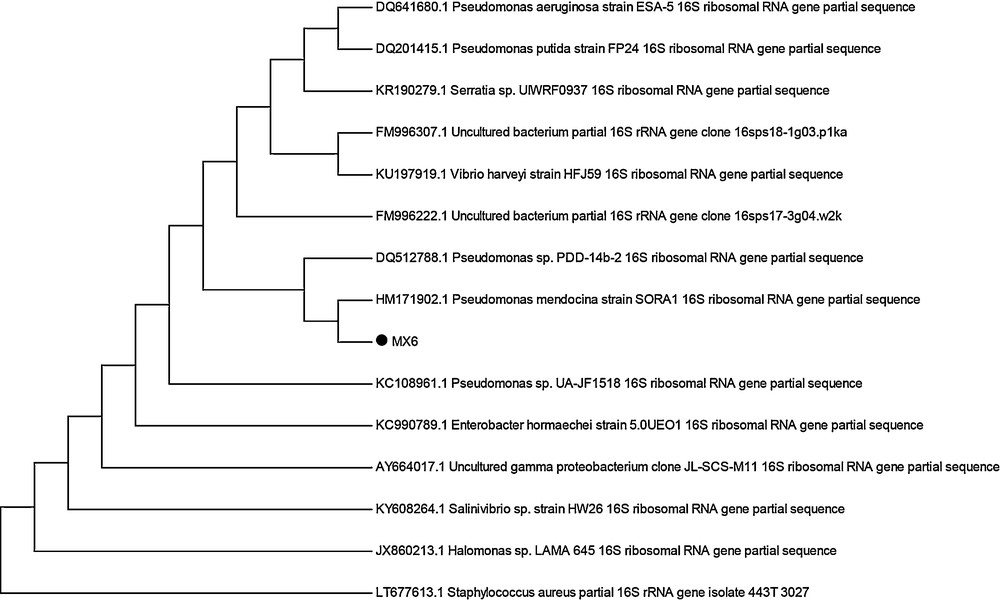
Neighbour-joining phylogenetic tree of bacterial isolate MX-6 and its closely matching homologues from NCBI BLAST results.
3.4 Determination of MIC
A high degree of resistance to all the selective metals was exhibited by MX-2 and Pseudomonas sp. MX-6. MIC values varied from 5.0 mM to 30 mM. Among the selected metals, Cr and Hg proved the most toxic ones. MIC of Hg and Cr for MX-2 and Pseudomonas sp. MX-6 were 5.0 mM and 2.5 mM, respectively. The least toxic metals for MX-2 were Co and Cu with an MIC value of 30 mM, while the least toxic metal for Pseudomonas sp. MX-6 was Zn as it resisted Zn up to 30 mM (Fig. 2).
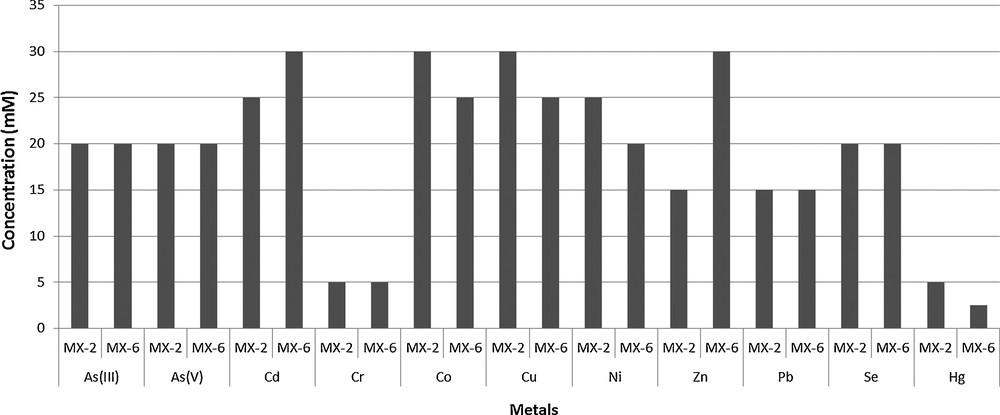
Minimum inhibitory concentrations of different metals against bacterial isolates.
3.5 Antibiotic resistance by multi-metal-resistant bacteria
Bacterial isolate MX-2 was found to resist chloramphenicol (3.0 μg·mL−1), ampicillin (1.0 μg·mL−1), kanamycin (3.0 μg·mL−1), and erythromycin (1.5 μg·mL−1) while sensitive to tetracycline (3.0 μg·mL−1). On the other hand, Pseudomonas sp. MX-6 was found resistant to chloramphenicol (3.0 μg·mL−1), but sensitive to all other antibiotics tested.
3.6 Oxidation/reduction of As by bacterial isolates
Bacterial isolates were screened for their ability to oxidize/reduce As. The results showed that both isolates had the ability to reduce As(V) into As(III) as well as to oxidize As(III) into As(V). The best reduction potential was exhibited by isolate Pseudomonas sp. MX-6. After 5 h of incubation in a normal saline solution, Pseudomonas sp. MX-6 reduced 506 μM As(V). Isolate MX-2 showed As(V) reduction up to 484 μM (Fig. 3).
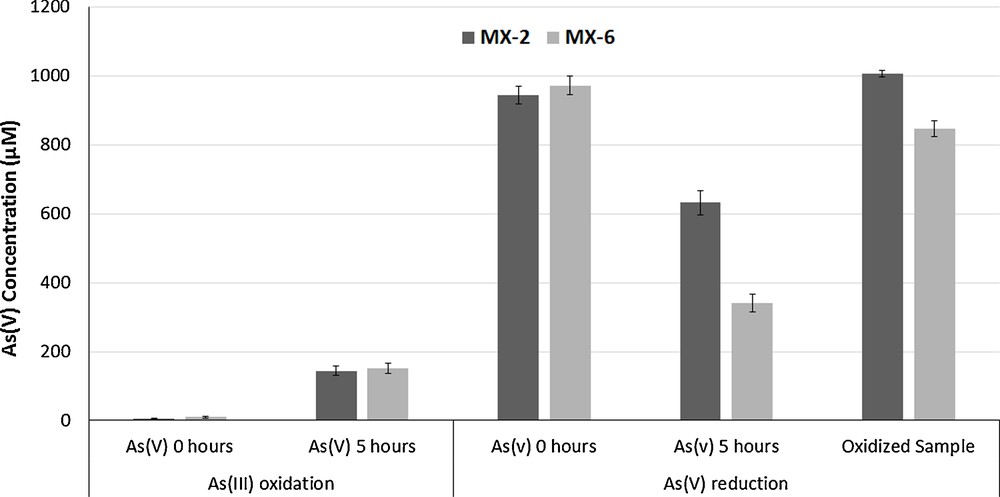
As(III) oxidation and As(V) reduction potentials of the bacterial isolates.
The As(III) oxidation potential of the isolates was also determined. After 5 h of incubation, it was found that best oxidation ability was exhibited by the isolate Pseudomonas sp. MX-6 (160 μM). On the other hand, MX-2 oxidized 150 μM As(III) into As(V) (Fig. 3).
3.7 Analysis of plasmids for metal-resistant genes
Plasmids were found to be present in both the isolates. The isolated plasmids were used to transform DH5α cells. Non-transformed DH5α was unable to resist all metals at a concentration of 5 mM and was sensitive to kanamycin and ampicillin. Thus, these two criteria were used for the screening of the transformants. In MX-2, resistance genes for As(III), As(V), Ni and Zn were found to be present on plasmids, as indicated by the transformation, while in Pseudomonas sp. MX-6 the genes for As(V), Cd, Se and Zn resistance were plasmid borne. We also analysed the resistance to antibiotics of the transformants, and it was observed that kanamycin- and ampicillin-sensitive DH5α was able to resist these two antibiotics after transformation. Pseudomonas sp. MX-6 plasmid conferred resistance against ampicillin as the corresponding DH5α transformants were able to resist ampicillin. Similarly, MX-2 plasmid was found to carry kanamycin resistance genes as kanamycin resistance was observed in the subsequent transformants. Non-transformed competent cells plated on LB-agar plates containing metals showed no growth, while colonies were observed on LB-agar plates not containing any metals.
3.8 Plant growth promoting characteristics of the isolates
Both isolates MX-2 and Pseudomonas sp. MX-6 were able to produce IAA and HCN. Maximum concentration of auxin (49.37 μg·mL−1) were produced by Pseudomonas sp. MX 6, while MX-2 produced auxin at a concentration of 34.06 μg·mL−1. MX-2 also showed phosphate solubilisation ability as clear zones were observed around the streaked lines.
3.9 Effects of isolates on plant growth
Shoot and root lengths, wet and dry weights, and the number of leaves of the plants were decreased in the presence of metals as compared to control plants. The shoot length of the plants was highly affected in the presence of As(V), As(III), Cr, Hg, and Pb. The mean shoot length decreased from 15.9 cm (control plants) to 11.33, 9.0, 11, 10.34 and 10.3 cm in the presence of As(V), As(III), Cr, Hg and Pb, respectively. The presence of MX-2 and Pseudomonas sp. MX-6 increased the shoot length of the plants to 19 and 19.2 cm, respectively. The shoot length was also increased by bacteria in the presence of metals as well, and most prominent results were shown by MX-2 in the presence of Hg (25 cm) and Pseudomonas sp. MX-6 in the presence of Se (23 cm) (Fig. 4).
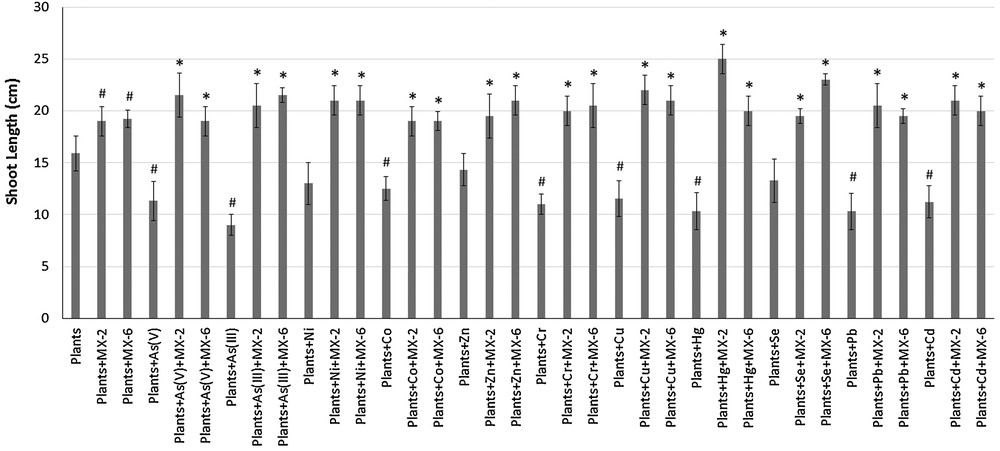
Effects of bacteria and metals on plant's (Vigna radiata) shoot length. #Significantly different from control plants, *Significantly different from plants + respective metal, P < 0.05.
The root length of the plants was most affected in the presence of As(V), As(III), Co, Cr and Hg. The mean root length of the control plants was 1.57 cm; it increased to 3 and 2 cm in the presence of MX-2 and Pseudomonas sp. MX-6, respectively. The root length decreased to 0.85, 0.65, 0.75, 0.72 and 0.5 cm in the presence of As(V), As(III), Co, Cr, and Hg, respectively. However, the presence of bacteria increased root length, even in the presence of metals, and most prominent results were shown by MX-2 in the presence of Zn (3.3 cm) and Pseudomonas sp. MX-6 in the presence of Se (3.1 cm) (Fig. 5). Moreover, an increase in the number of leaves as well as an increase in wet and dry weights was also observed in the presence of the bacteria (Table 1).

Effects of bacteria and metals on plant's (Vigna radiata) root length. #Significantly different from control plants, *Significantly different from plants + respective metal, P < 0.05.
Effects of the bacterial isolates on the number of leaves, wet and dry weight of Vigna radiata.
| Number of Leaves | Wet Weight | Dry Weight | ||||||||||
| Conditions\Isolates | Mx-2 | SD ± | MX-6 | SD ± | Mx-2 | SD ± | MX-6 | SD ± | Mx-2 | SD ± | MX-6 | SD ± |
| Control Plants | 4.2 | 0.9 | 4.2 | 0.9 | 0.315 | 0.03 | 0.315 | 0.03 | 0.04 | 0.006 | 0.04 | 0.006 |
| Plants + isolate | 4.4 | 0.54 | 4.8 | 0.54 | 0.36 | 0.038079 | 0.35 | 0.044159 | 0.025 | 0.0055 | 0.045 | 0.003 |
| Plants + As(V) | 2.666667 | 0.4 | 2.666667 | 0.4 | 0.24 | 0.03 | 0.24 | 0.03 | 0.03 | 0.004 | 0.03 | 0.004 |
| Plants + As(V) + isolate | 3.33 | 0.4 | 5.333333 | 0.6 | 0.33 | 0.014142 | 0.265 | 0.014142 | 0.034 | 0.005 | 0.035 | 0.005 |
| Plants + As(III) | 3 | 0.23 | 3 | 0.23 | 0.18 | 0.02 | 0.18 | 0.02 | 0.028 | 0.0056 | 0.028 | 0.0056 |
| Plants + As(III) + isolate | 4.333333 | 0.57735 | 6 | 0.4 | 0.26 | 0.014142 | 0.22 | 0.014142 | 0.032 | 0.0065 | 0.033 | 0.004 |
| Plants + Ni | 3.333333 | 0.33 | 3.333333 | 0.33 | 0.25 | 0.03 | 0.25 | 0.03 | 0.037 | 0.005774 | 0.037 | 0.005774 |
| Plants + Ni + isolate | 5.333333 | 0.55 | 5.333333 | 0.76 | 0.27 | 0.014142 | 0.26 | 0.014142 | 0.03 | 0.001 | 0.041 | 0.004 |
| Plants + Co | 4 | 0.45 | 4 | 0.45 | 0.27 | 0.024 | 0.27 | 0.024 | 0.023333 | 0.005774 | 0.023333 | 0.005774 |
| Plants + Co + isolate | 4.666667 | 0.76 | 5.333333 | 0.34 | 0.31 | 0.014142 | 0.31 | 0.014142 | 0.03 | 0.006 | 0.025 | 0.0025 |
| Plants + Zn | 4 | 0.46 | 4 | 0.46 | 0.27 | 0.020817 | 0.27 | 0.020817 | 0.031 | 0.0036 | 0.031 | 0.0036 |
| Plants + Zn + isolate | 6 | 0.46 | 6.666667 | 0.35 | 0.29 | 0.014142 | 0.3 | 0.014142 | 0.04 | 0.004 | 0.035 | 0.004 |
| Plants + Cr | 2.666 | 0.13 | 2.666 | 0.13 | 0.21 | 0.026 | 0.21 | 0.026 | 0.021 | 0.0016 | 0.021 | 0.0016 |
| Plants + Cr + isolate | 4.666667 | 0.44 | 4.66 | 0.65 | 0.24 | 0.014142 | 0.36 | 0.014142 | 0.026 | 0.005 | 0.03 | 0.0015 |
| Plants + Cu | 3.333333 | 0.57735 | 3.333333 | 0.57735 | 0.25 | 0.030551 | 0.25 | 0.030551 | 0.026 | 0.0035 | 0.026 | 0.0035 |
| Plants + Cu + isolate | 4.666667 | 0.66 | 6 | 0.222 | 0.3 | 0.014142 | 0.32 | 0.014142 | 0.03 | 0.0025 | 0.027 | 0.0025 |
| Plants + Hg | 2.66 | 0.32 | 2.66 | 0.32 | 0.19 | 0.026 | 0.19 | 0.026 | 0.026 | 0.0025 | 0.026 | 0.0025 |
| Plants + Hg + isolate | 5 | 0.44 | 6 | 0.444 | 0.24 | 0.014142 | 0.23 | 0.014142 | 0.029 | 0.003 | 0.03 | 0.003 |
| Plants + Se | 2.666667 | 0.6 | 2.666667 | 0.6 | 0.216667 | 0.015275 | 0.216667 | 0.015275 | 0.03 | 0.0025 | 0.03 | 0.0025 |
| Plants + Se + isolate | 4 | 0.5 | 5.333333 | 0.66 | 0.265 | 0.014142 | 0.35 | 0.044159 | 0.034 | 0.003 | 0.037 | 0.003 |
| Plants + Pb | 2.666667 | 0.42 | 2.666667 | 0.42 | 0.245 | 0.035 | 0.245 | 0.035 | 0.028 | 0.005774 | 0.028 | 0.005774 |
| Plants + Pb + isolate | 4.666667 | 0.11 | 6.666667 | 0.23 | 0.25 | 0.014142 | 0.254 | 0.014142 | 0.031 | 0.0051 | 0.031 | 0.0051 |
| Plants + Cd | 4 | 0.6 | 4 | 0.6 | 0.24 | 0.01 | 0.24 | 0.01 | 0.025 | 0.004 | 0.025 | 0.004 |
| Plants + Cd + isoalte | 4 | 0.44 | 5.333333 | 0.75 | 0.27 | 0.014142 | 0.265 | 0.014142 | 0.03 | 0.002 | 0.031 | 0.0022 |
4 Discussion
Most of the industries in developing and under-developing countries release their untreated wastes directly into the environment, resulting in noxious concentrations of As and of other toxic metals and metalloids. Soil samples were collected from wastewater treatment plant, Kasur, Pakistan, which receives both industrial and domestic wastewater. Microorganisms that survive in such contaminated environments have developed strategies to resist and detoxify contaminants. Direct spreading of soil dilution on media plates without any metal resulted in a large diversity of bacteria, but yielded only trivial number of bacterial colonies on TSA media containing As(III), As(V), Cr, Cd, Co, Hg, Zn, Ni, Pb and Se. However, when an enrichment culture technique was used, a significant number of bacterial colonies were obtained, clearly indicating decreased microbial load in such heavily polluted environment. In the present study, multi-metal-resistant bacteria were also found to resist many antibiotics. As genetic determinants for metals and antibiotics are mostly present on plasmids, it may, therefore, be considered that the incidence of antibiotic resistance observed in these metal-resistant bacteria was due to the increased selection pressure at their habitat. Co-relation of antibiotic and metal resistance has already been reported and it was found that the functional and structural characteristics of metal and antibiotic resistance shared common themes [21].
The ability of the isolates to oxidize/reduce As was also determined. Oxidation and reduction are the two most commonly employed mechanisms by bacteria to resist and detoxify metals. Bacterial isolates were found to perform As-oxidation as well as reduction. Bacteria are known to either oxidize or reduce As [22–24]. However, there are a few rare recent studies that report bacteria capable of both reducing as well as oxidizing As [11,25]. This peculiar characteristic could offer novel insights into bacterial mechanisms of As-detoxification [1]. In contaminated soils, bacteria possessing dual ability of As(V) reduction and As(III) oxidization provide advantage in reducing As-toxicity. Moreover, this mechanism serves to maintain the biogeochemical cycle of different species of As in microenvironments [25].
The transformation of DH5α, sensitive to metals, with plasmids isolated from multi-metal-resistant bacteria yielded transformants having the ability to resist many metals. It was observed that MX-1 plasmid conferred resistance to As(III), As(V), Cr, Cd, N, Se, and Hg, while MX-2 plasmid transformants were resistant against As(III), As(V), Cr, Zn and Ni. Transformants of Pseudomonas sp. MX-6 plasmid showed resistance towards As(V), Zn, Se, and Cd. This indicated that these resistant genes were plasmid borne. Plasmid-encoded genes represent a substantial pool of mobile DNA that can contribute to the genetic adaptation of microbial communities. The number of plasmid carrying bacteria are two-ten folds higher in polluted environments as compared to non-polluted sites [26]. Therefore, the probability of metal-resistant bacteria to carry plasmids is much higher as compared to bacterial communities of pristine environments [27]. The transformation of DH5α with plasmid isolated from metal-resistant bacteria is well documented [28,29]. Co-transfer of antibiotic and metal resistance genes has also been previously reported [30]. The bacteria also enhanced the growth of Vigna radiata (mung bean plant). A significant increase in root lengths and shoot lengths, wet and dry weights as well as number of shoots and leaves was observed both in the absence and presence of metals. Bacteria not only carried plant growth promoting traits such as auxin production, phosphate solubilisation, etc., but were also capable to resist the metals (and detoxify them as well). This provided extra benefit to the plants, which, therefore, showed better growth in the presence of bacteria, even in the presence of the metals. Auxin is a well-known plant growth promoting substance, and its presence is known to enhance plant growth [31]. Phosphate is another important plant growth promoting factor, and phosphate-solubilizing bacteria are known to enhance plant growth by solubilizing phosphates, which are then easily taken up by the plants [32].
5 Conclusion
The bacterial isolates were able to resist multiple metals and were also capable of redox transformation of As. The bacteria also enhanced plant growth both in the absence and presence of metals. Therefore, it can be concluded that bacteria with such diverse abilities can help in the development of effective bioremediation processes and reclamation of contaminated lands for agriculture purposes. Such bacteria can be used as biofertilizers to enhance crops production even in metal-contaminated soils.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
University of the Punjab, Lahore, is acknowledged for funding to carry out this research.


