1 Introduction
Metal-contaminated soils pose a severe threat to environment, agriculture, and consequently via the food chain to human and animal health [1].
Cadmium is one very prevalent heavy metal and is one of the most toxic pollutants of the surface soil layer. It is introduced into soils by synthetic fertilizers and pesticides, widespread due to mining, smelting and power station industry activities [2]. This metal is non-essential, but poisonous for plants, animals, and humans [3]. In plants, cadmium inhibits shoot and root growth and affects nutrient uptake, homeostasis and physiological processes such as photosynthesis [4].
This is why cadmium pollution attracts the most attention of environmentalists around the world. Like other heavy metals, cadmium affects the qualitative and quantitative structure of microbial communities, resulting in decreased metabolic activity and diversity [5]. Remediation of soils contaminated by heavy metals can be done using chemical, physical, and biological techniques [6]. Chemical and physical methods are expensive and cause secondary pollutants [7]. Phytoremediation is the biological technique which refers to the use of some plants for bioremediation. Considerable amounts of toxic metals can be accumulated through bioremediation, and it can play a significant role in cleaning up metal pollutants [8]. Recently, legumes associated with plant-growth-promoting rhizobacteria have been extended to bioremediation of both organic and metal pollutants [9–11]. PGPRs have the exceptional ability to promote the growth of plants by different mechanisms such as nitrogen fixation, solubilisation of minerals, transformation of nutrient elements, production of phytohormones, and sequestration of iron by siderophores [12]. In addition, PGPRs had different heavy metal tolerance mechanisms; they involve biosorption, precipitation or bioaccumulation in external and intracellular spaces. They contribute in the accumulation of heavy metals in forage plants especially in roots and minimize their translocation for shoots [11].
The aim of this work was to identify a cadmium-tolerant soil bacteria isolated from nodules of Sulla coronaria cultivated in heavy metal contaminated soil samples and to form a symbiotic system useful for the phytostabilization of Cd-contaminated soils, being known that this plant is one of the most important for fodder; it is cultivated in several countries of the western Mediterranean basin, especially in Spain, Italy and North Africa [13,14]. It is a species well-adapted to a large range of environmental conditions and actually shows a marked resistance to extreme conditions of drought, salinity, and alkaline pHs as high as 9.3 [15].
2 Materials and methods
2.1 Determination of toxic metals in soil samples
Four composite soils were collected from the north of Tunisia: S1 Jerissa (35°51′N, 8°38′E), which is an agricultural site not contaminated by heavy metals and used as a control. S2 Ghezela (37°05′02″N, 9°32′08″E), S3 Manzel Bourguiba (37°9′N, 9°47′E) located in Bizerte governate and near a large iron mine and S4 Jebal Ressas (36°36′N, 10° 20′E) is an agricultural area located near an old lead–zinc (Pb–Zn) mine. Three samples were taken at a depth of 15 cm from the soil surface and separated by 10 m. The collected soil samples were air dried at 40 °C for 16 h and sieved into coarse and fine fractions. Total soil heavy metals were extracted by dissolving 0.5 g of soil sample in 6 mL of hydrochloric acid and 2 mL of 2% nitric acid at 95 °C for 75 min, filtered and then diluted to 50 mL with distilled water. Extractable heavy metals were extracted with 1 M NH4OAc [16]. The above heavy metal concentrations in the extracts were determined by inducted coupled plasma optical spectrophotometer (ICP-OES) after aqua regia treatment in a microwave system.
The soil pH was measured in distilled water with a soil/solution ratio of 1:2.5 (W:V). The organic matter content and the total phosphorus in the soil were determined according to the methods described in [16].
2.2 Isolation of strains and culture conditions
Seeds of Sulla coronaria Bikra 21 were obtained from UTAP (« Union tunisienne de l’agriculture et de la pêche ») station of Manouba, located to the northwest of Tunis. Its surface was sterilized with 70% ethanol for 1 min then rinsed five times with distilled water. Seeds were germinated on 9 ‰ agar plates at 28 °C in darkness and grown in pots of 1 kg of soil from the contaminated sites (3 plants/pot) and 10 pots from each soil were placed in a controlled plant-growth chamber (16 h light, 8 h dark/relative humidity 60%) and watered with distilled water. The experiment has been performed for 2 months.
The collected nodules of Sulla Coronaria were washed with sterile water and then surface sterilized using 70% ethanol and 0.2% HgCl2, then thoroughly washed with sterile distilled water. After surface sterilization, the nodules were crushed and then streaked in YEMA medium (Yeast Extract Mannitol Agar) [17]. Single colonies were selected and checked for purity by repeated streaking. All isolates were incubated at 28 °C and maintained in a 20% (v/v) glycerol solution at –80 °C.
2.3 Screening test for cadmium and heavy-metal-resistant strains
To isolate cadmium-resistant strains, the isolates were screened on YEMA plates, supplemented with different concentrations of Cd in the form of cadmium chloride (0.02, 0.06, 0.1, 0.4, 0.8, 1, 1.2, 1.6, 2, 2.4, 3, 4.1, 4.5 mM). Ni (in the form of Nickel chloride) was used at concentrations of 0.008, 0.016, 0.024, 0.06, 0.08, 0.12, 0.2 and 0.28 mM, Pb (in the form of lead chloride) was used at concentrations of 0.27, 0.675, 0.94, 1.35, 2.025, 2.7, 3.375, and 4.05 mM, Zn (in the form of zinc sulfate) was used at concentrations of 0.6, 0.8, 1, 1.2, 1.28, 1.36, 1.4 and 1.6 mM, and Cu (in the form of copper sulfate) was used at concentrations of 0.01, 0.02, 0.05, 0.075, 0.1, 0.15, 0.25, and 0.35 mM. Thus, the maximal concentration used for resistance testing was similar to those of the corresponding elements found in contaminated soils.
Multi-point inoculators were used to add a 20-μL aliquot of cell suspension from each sample isolate to the YEMA plates, and then the culture was incubated at 28 °C for 3–5 days. The minimum inhibitory concentration (MIC) was defined as the lowest concentration at which no viable colony-forming units (CFU) were observed after 3 days of incubation at 28 °C.
2.4 PCR amplification and RFLP of the 16S rRNA gene
Regarding DNA extraction, the method described by [18] was used. Primers fD1 5′-GGAGAGTTAGATCTTGGCTC-3′ and rD1 5′-AAGGAGGTGATCCAGCCGCA-3′ were used for the amplification of almost full length 16S rRNA genes, around 1530 base pairs per region using the Bio-RAD Thermal Cycler model (C1000).
PCR reactions were done in a final volume of 25 μL containing the following components: 2.5 μL of 10X reaction buffer (10 mM Tris-HCl, 50 mM MgCl2), 1 μM of DNA primer, 1.5 mM of MgCl2, 200 μM of dNTP, 1.5 U of Taq polymerase (Appligene 5U μL−1), 1.5 μl (10 ng) of bacteria DNA template.
DNA amplification was carried out as follows: 1 cycle at 94 °C for 3 min; 35 cycles at 94 °C for 50 s, 54 °C for 45 s, and 72 °C for 90 s; 1 cycle at 72 °C for 8 min.
DNA fragments resulting from PCR reactions were electrophoresed in 1% (w/v) agarose gels for 20 min and visualized by ethidium bromide staining.
Sixteen rDNA PCR/RFLP analyses were carried out by digesting the respective PCR products with the endonucleases MspI, NdeII. DNA fragments resulting from PCR reactions, treated with endonucleases, were electrophoresed in 3% (w/v) agarose gels for 3 h and visualized by ethidium bromide staining.
Species assignation was done according to 16S rDNA typing and comparison with the published database of mapped restriction sites in the 16S rRNA genes of rhizobia [18].
2.5 Nodulation assay
Inoculation and seed treatment were performed by using the method of Vincent [17]. The plants were cultured in a growth chamber at a constant temperature of 23 °C and a light/dark photoperiod of 16/8, hand watered with a nitrogen-free nutrient solution [17]. Six repetitions in six different pots were used for every strain, and sterile sand was used as the non-inoculated control (negative control).
2.6 Phylogenic analysis of the 16S rRNA gene
The 16S rRNA gene of the four selected strains was amplified with the same primers and procedure as in the restriction analysis, and was sequenced directly by using a PCR-based method. Nucleotide sequence similarity searches were conducted by Genbank BLAST (N).
2.7 Production of Indole Acetic Acid (IAA) and siderophores
Each bacterial strain selected was incubated in 10 mL of an SMS medium (sucrose, 10 g.L−1; [NH4]2SO4, 1 g.L−1; K2HPO4, 2 g.L−1; MgSO4·7H2O, 0.5 g.L−1; yeast extract, 0.5 g.L−1; CaCO3, 0.5 g.L−1; NaCl, 0.1 g.L−1 and pH 7.2), supplemented with 0.5 mg.mL−1 of tryptophane for 5 days at 28 °C with shaking at 150 rpm. Cells and supernatants were separated by centrifugation at 10,000 g for 5 min and 1 mL of supernatant was mixed with 100 μL of orthophosphoric acid (10 mM) and 2 mL of Salkowsky reagent (2% 0.5 M FeCl3 in 35% perchloric acid) and incubated at 28 ± 2 °C in darkness for 30 min. The absorbance of the pink color developed was read at 530 nm. The IAA concentration in the supernatant was determined using a standard IAA solution [19]. The experiments were repeated three times on different time intervals. Siderophores production was detected using Chrome Azurol S (CAS) plates [20].
2.8 Phosphate solubilisation
Plates containing Trypticase Soya Agar medium supplemented with Ca5(PO4)3OH were inoculated with 20 μL of a LB pure bacterial culture. Plates were incubated at 28 °C and observed daily during 7 days until the formation of transparent “halos” around each colony [10].
2.9 Bioaccumulation of Cd in bacteria
The cells (0.1 g of dry weight, DW) were inoculated into 100 mL of a TY medium containing 0.1 mM Cd2+ and incubated at 28 °C with agitation at 150 rpm. The cells were harvested at 24 h and 48 h after inoculation by centrifugation at 6000 g for 25 min. After washing three times with sterilized ddH2O, the cell pellets were agitated with 10 mM of sterilized EDTA at 28 °C at 150 rpm for 10 min in order to remove the cadmium ions adsorbed on the cell surface. After further centrifugation at 14,000 g for 25 min, the cells were suspended in 5 mL of 0.1 M HNO3, and the solution was centrifuged at 14,000 g for 30 min. The supernatant was used for the determination of cell wall-bound Cd2+ with Flame Atomic Absorption Spectrometry. After measurement of the dry weights, the cell pellets were digested with H2SO4–HClO4 (V:V = 3:1) at 110 °C for 3 h and diluted with ddH2O, for the determination of intracellular accumulated Cd2+ [21].
2.10 Amplification of Cd-resistant gene partial sequence
The strains were incubated in TY medium, and the total DNA was extracted using the method reported by [22]. Four primers: P1: Cad A1 forward (GAAAGAAGATAAAGTGCCGTTTT), Cad A1 reverse (ATGCACAAGGACAACCAACA), P2: Cad A3 forward (TGAAAAATCTGCTCGCCAAG), Cad A3 reverse (ATGCACAAGGACAACCAACA), P3: Cad A2 forward (CTCGCCAAGCTTCACAAGAG), Cad A2 reverse (ATGCACAAGGACAACCAACA), P4: CZC1 forward (AACCAGATCTCGCGCGAGAAC), CZC1 reverse (CGGCAACACCAGTAGGGTCAG) for the amplification of Cd-resistant genes were utilized. The conditions for PCR amplification were: pre-denaturalization at 95 °C for 3 min, then 30 cycles of denaturing at 94 °C for 1 min, annealing for 1 min for extension at 50 °C (P2), 52 °C (P1, P3) and 55 °C (P4) for 2 min, and a final step for extension at 72 °C for 10 min. The PCR product was detected as described by [23].
2.11 Statistical analysis
All experiments were done with at least three replications and repeated independently. The average and standard deviations were calculated (±SD) by Microsoft Excel 2007 and Statistica 6.0
3 Results
3.1 Determination of toxic elements in soil samples
In the S1 soil, all concentrations of heavy metals were under the French norm for agricultural soil [24]. The S2 soil samples were severely contaminated by Pb (10,100 mg.kgDW−1) and Cd (16.5 mg.kgDW−1), and their levels were higher than those defined by the French norm (Table 1). The S3 soil sample was mainly characterized by its Pb contamination (426 mg.kgDW−1). In the S4 soil sample, Pb contamination was moderate (116 mg.kgDW−1), but higher than those defined by the French norm. This site was richer in organic matter and phosphorus; it is currently used as an agricultural zone for viticulture.
Characteristics and heavy metal concentration analyzed in the soils used in the experiment; S1: Jerissa, S2: Ghzela, S3: Manzel Bourguiba, S4: Jbel Ressas. Results are in mg.kg–1. FN: French norms for agricultural soils.
| Heavy metals concentration (mg.kgDW–1) | S1 | S2 | S3 | S4 | FN |
| Cd total | 0.27 | 16.5 | 0.27 | 0.841 | 0.7–2 |
| Cd extractible | < 0.27 | < 0.27 | < 0.27 | < 0.27 | |
| Cu total | 10.6 | 21.6 | 20.0 | 40.3 | 35–100 |
| Cu extractible | < 0.97 | < 0.97 | < 0.97 | < 0.97 | |
| Pb total | 27.2 | 10100 | 426 | 116 | 60–100 |
| Pb extractible | < 1 | 143 | 50 | < 1 | |
| Zn total | 31.2 | 235 | 84.5 | 211 | 150–300 |
| Zn extractible | – | 121 | – | 48 | |
| Soil pH | 8.1 | 9.04 | 7.5 | 8.23 | 5.5–7.5 |
| % Carbon | 0.55 | 0.65 | 1.1 | 2.32 | No limit |
| Phosphorus (P2O5, ppm) | 21 | 16.5 | 31 | 541.7 | 70–200 |
| %Organic matter | 1.5 ± 0.3 | 1.11 ± 0.6 | 1.15 ± 0.2 | 4.02 ± 0.6 | No limit |
| ‰ n total | 1.87 | 0.33 | 1.68 | 2.37 | |
| Potassium (K2O, ppm) | 822 | 84 | 877.2 | 878.4 |
3.2 Tolerance of the isolates to heavy metals
A total of 82 strains were isolated from the nodules of the Sulla coronaria grown in the four soil samples. The phenotypic characterization of the isolates for tolerance to heavy metals was done by measuring the MIC of the strains (Table 2). Our study showed that 73 isolates tolerated 4 mM of Pb and 48 resisted to 1.6 mM of Zn. The majority of the isolates were sensitive to Cd and Cu: only 21 of them could grow in a medium supplemented with 2.6–4.1 mM of Cd and 34 isolates tolerated 0.25–0.35 mM of Cu. Nickel was especially lethal and only 16 were resistant to 0.12–0.2 mM of Ni. Some strains were particularly resistant to several elements at the same time (Table 2).
Tolerance to heavy metals (Pb, Zn, Cu, Ni and Cd) of isolates isolated from nodules of Sulla Coronaria.
| Heavy metals | ||||||
| Pb | Zn | Cu | Ni | Cd | ||
| Sensible | Concentration (mM) | (0.2–1) | (0.6–1) | (0.01–0.07) | (0.008–0.032) | (0.01–1) |
| Strains number | 4 | 6 | 27 | 19 | 47 | |
| Tolerant | Concentration (mM) | (1.3–2.7) | (1.2–1.3) | (0.1–0.2) | (0.04–0.08) | (1.1–2.5) |
| Strains number | 5 | 28 | 21 | 47 | 14 | |
| Very tolerant | Concentration (mM) | (2.8–4) | (1.3–1.6) | (0.25–0.35) | (0.12–0.2) | (2.6–4.1) |
| Strains number | 73 | 48 | 34 | 16 | 21 |
3.3 Molecular diversity of the strains
The 16S rDNA region of the 82 isolates, selected from Sulla coronaria, was amplified by PCR. Electrophoresis revealed from one to five profiles for each enzyme used. The number of bands varied from 70 bp to 690 bp for Ndell (Fig. 1). Despite the high diversity, restriction enzymes produced polymorphic banding patterns, the most discriminative of which was that obtained with Mspl.

PCR/RFLP profiles of 16S rRNA genes of 9 isolates (I1…I9) digested with (A) MspI and (B) with NdeII and separated by electrophoresis; Mq: molecular marker (100 pb).
The collection was represented by different bacterial genera. Thirty-four percent of the isolates were related to Rhizobium sp. The genus Agrobacterium was represented by 8%, whereas only 2% of isolates belonged to the genus Sinorhizobium. Finally, PCR-RFLP of the 16S rDNA gene could not discriminate 56% of the isolates, which were not identified.
3.4 Plant nodulation test and symbiotic effectiveness
Among the 82 isolates, 19 were selected and tested for their ability to nodulate Sulla coronaria. These strains represent the different profiles of digestion obtained by PCR/RFLP (Table 3). The nodulation test showed that only 9 strains nodulated the host plant.
Growth and nodulation test of Sulla coronaria inoculated by the 19 isolates (SDW: shoot dry weight; RDW: root dry weight; Nod·plant–1: nodule per plant). The values are the average of three repetitions in three independent pots) and the molecular characterization of 19 isolates by PCR/RFLP of the 16rDNA gene.
| Locality | Nod·plant–1 | % SDW | % RDW | PCR/RFLP characterization | |
| Control | – | 0 | 100 | 100 | – |
| I1 | S4 | 0 | 77.01 | 94.94 | Agrobacterium |
| I2 | S1 | 0 | 73.39 | 78.4 | Agrobacterium |
| I3 | S2 | 4 | 168.7 | 72.18 | No identified |
| I4 | S2 | 3 | 104.03 | 106.5 | R. leguminosarum |
| I5 | S2 | 0 | 112.4 | 97.07 | Rhizobium sp. |
| I6 | S3 | 4 | 139.2 | 103.95 | R. leguminosarum |
| I7 | S2 | 0 | 79 | 57.68 | No identified |
| I8 | S2 | 15 | 153.8 | 150 | Rhizobium sp. |
| I9 | S2 | 3 | 173.33 | 151.39 | No identified |
| I10 | S2 | 3 | 162.1 | 89.09 | No identified |
| I11 | S3 | 0 | 104.33 | 59 | S. melliloti |
| I12 | S1 | 7 | 131.21 | 76.35 | No identified |
| I13 | S1 | 0 | 91.98 | 54.39 | R. phaseoli |
| I14 | S2 | 0 | 81.65 | 71.52 | Agrobacterium |
| I15 | S1 | 0 | 124.22 | 85.79 | No identified |
| I16 | S2 | 5 | 110.33 | 92 | Rhizobium sp. |
| I17 | S1 | 0 | 71.19 | 52.48 | Rhizobium sp. |
| I18 | S2 | 0 | 126.48 | 86 | R.leguminosarum |
| I19 | S1 | 2 | 53.44 | 48.17 | Agrobacterium |
Inoculation of Sulla coronaria by I8 and I9 enhanced SDW by 153% and 173%, respectively, and RDW increased up to 150 and 151%, respectively. In addition, inoculation with I3 and I10 improved SDW by 169% and 162%, respectively, whereas inoculation with the strain I19 conferred the lowest growth to Sulla coronaria (Table 3).
3.5 Selection of the inoculum
Taking into consideration the isolates tolerance to Cd and nodulation capacity, four strains were chosen. The I3 strain had the lowest MIC (0.25 mM Cd), but it improved plant growth. The I8 and I9 strains tolerated 1 mM Cd, enhanced the SDW of plants, and it was noted that I8 gave the largest number of nodules per plant. I3 and I9 had little white nodules, and I8 strains had red nodules. Among the selected strains, I10 was considered the most tolerant to Cd, due to its resistance up to 4.1 mM.
The nucleotide sequence of the amplified DNA was determined and compared with available 16S rRNA sequences to construct a phylogenetic tree. The subsequent BLAST analysis in GenBank affirmed that the strain I9 have 96% homology with pseudomonas fluorescens strain MFN 1032, I10 showed 88% similarity with uncultured pseudomonas sp., clone A6B-D10, I3 have 96% homology with Rhizobium sullae strain IS123 and I8 has 97% similarity with Rhizobium sullae IS 123.
3.6 Production of indole acetic acid (IAA), siderophores and solubilization of P
Our results showed that I8 (Rhizobium sullae) showed a higher production of IAA (64.18 μg.μL−1) compared to the rest of the studied bacteria, whereas the production of siderophores was positive in I3, I8 and I10 (Table 4).
Indole acid acetic (IAA), siderophores production and P solubilization of isolates. The values are the average ± SD of three repetitions.
| Strain | CMI (mM Cd) | IAA (μg/μL) | Siderophores | Solubilization of P |
| I3 (Rhizobium sullae) | 0.25 | 21.58 ± 1.35 | + | – |
| I8 (Rhizobium sullae) | 1 | 64.18 ± 0.35 | + | – |
| I9 (P. fluorescens) | 1 | 2.30 ± 0.11 | – | + |
| I10 (Pseudomonas sp.) | 4.1 | 0.68 ± 0.15 | + | + |
The isolates I3 and I8 were not able to solubilize the P contrarily to I9 and I10 (Table 4).
3.7 Estimation of Cd bioaccumulation in the screened bacteria
The accumulation of cadmium varied between cells sites and incubation times; in addition it increased with Cd tolerance. The highest cadmium accumulation appeared in the whole cells of I10 isolates at 48 h, reaching 14737.54 μg.gDW−1 (Table 5). The Concentration of cadmium in cell walls were the lowest in I3 at 24 h (0. 57 μg.gDW−1), therefore it enhanced significantly at 48 h (23 μg.gDW−1). The highest level was in I10 at 24 h (70 μg.gDW−1), but it declined by 65% at 48 h, whereas it was enhanced by 67% in I9. The intracellular Cd accumulation were enhanced by increasing the time of incubation in the four strains (Table 5); the maximum content was obtained at 48 h in I10 (14513 μg.gDW−1).
Cadmium accumulations in two compartments of isolates: cell and cell wall in (μg.g.DW–1) at two times, 24 and 48 h. The values are the average ± SD of three repetitions.
| Strain | Intracellular Cd | Cell wall Cd | ||
| 24 h | 48 h | 24 h | 48 h | |
| I3 (Rhizobium sullae) | 7035 ± 34 | 7844 ± 25 | 0.57 ± 0.1 | 23.44 ± 3 |
| I8 (Rizobium sullae) | 387 ± 18 | 3703 ± 35 | 37.64 ± 4 | 33.47 ± 5 |
| I9 (P. fluorescens) | 2.56 ± 1 | 2118 ± 47 | 19.9 ± 3 | 33.34 ± 6 |
| I10 (Pseudomonas sp.) | 264 ± 23 | 14713 ± 65 | 70 ± 12 | 24.54 ± 3 |
3.8 Detection of resistance genes in the screened bacteria
In the four selected soil bacteria, four primers were selected and identified by primer Blast from NCBI. The amplification of the Cd-resistant gene showed different bands with different size (Table 6). The alignment of Cd-resistant gene PCR showed a high homology with Cd-resistant genes from other bacteria. In I3 (Rhizobium sullae), a band of 1200 bp was found with primer1, which had 90% homology with Pseudomonas fluorescens.
PCR amplification of Cd-resistant genes using different primers.
| Strain | Primer1 | Primer2 | Primer3 | Primer4 |
| I3 | 1200 pb | – | – | – |
| I8 | 1600 pb | – | – | – |
| I9 | – | 500 pb | 1200 pb | 500 pb |
| I10 | 1300–600 pb | 1300 pb | – | – |
In I8 (Rhizobium sullae), we evidenced a band of 1600 pb with primer 1, the comparison of the Cd gene sequence indicated 85% homology with Sinorhizobium meliloti plasmid pHRC017.
In I9, one band was revealed with each tested primers (P2:500 pb; P3: 1200 pb and P4: 500 pb, Fig. 2) and the sequence analysis showed homology with Pseudomonas fluorescens by 90%. In I10, a band of 1300/600 pb was found with primer1 and a band of 1300 pb with primer 2 (Fig. 2); these sequence showed 99% similarity with Pseudomonas putida (Table 6).
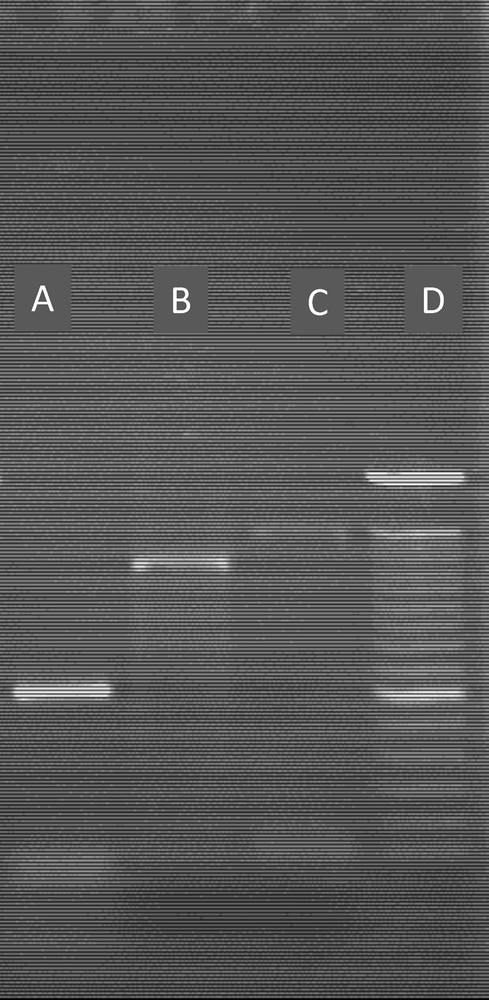
PCR Amplification of a Cd-resistant gene in the two selected bacteria (I9 and I10) with different primers. A: I9with Primer P4; B: I9 with primer P3; C: I100 with primer P2; D: molecular marker (100 pb).
4 Discussion
4.1 Tolerance of the isolates to heavy metals
In the present study, we have obtained a collection of soil bacteria isolated from nodules of Sulla coronaria cultivated in soils contaminated by heavy metals, which even influence the growth, abundance, tolerance, qualitative and quantitative structure of microbial communities [25]; it is well known that the bacteria isolated from polluted soil are resistant to higher concentrations of heavy metals than those isolated from unpolluted area [26]. The Pb-tolerance study of soil bacteria indicated that the majority of isolates tolerated 2.8–4 mM Pb; these values were comparable to Pb resistant PGPRs isolated from Lens culinaris cultivated in Pb contaminated soils [27], whereas Bradyrhizobium and Sinorhizobium can grow in media containing up to 2 mM Pb [11]. The Ni and Zn tolerance of isolates was lower than that of Rhizobium leguminosarum isolated from a sewage-sludge-treated soil and having a tolerance of 0.24 mM Ni and 6–8 mM Zn [28]; elsewhere, the tested soil bacteria were sensitive to Cu comparatively to others such as Agrobacterium tumefaciens [23] and Sinorhizobium meliloti [21], which can tolerate up to 2 mM Cu. The level of resistance to Cd in these bacteria is comparable to those of other bacteria from different genera such as Azorhizobium caulinodans, Staphylococcus aureus, Bacillus subtillis, and E. coli [29].
Even though metals exert their toxic effects on the diversity of microorganisms [1], the most Cd-resistant soil bacteria were isolated from nodules of Sulla coronaria cultivated in Cd-contaminated soil S2, heavy metal tolerant bacteria could survive in these sites and were selected for their potential application in the phytoremediation of contaminated soils [25].
4.2 Molecular diversity and symbiotic effectiveness of heavy-metal-resistant strains
The molecular study of strains isolated from nodules of Sulla coronaria showed a large diversity represented essentially by Rhizobium sp., R. leguminosarum, Agrobacterium sp., Pseudomona sp., and Rhizobium sullae. The genus Sinorhizobium sp. was the least represented. In many studies, heavy-metal-contaminated soils can serve as a source for the isolation of heavy-metal-resistant bacteria [21,30] and Rhizobium can be used as an organism indicating contamination by several heavy metals as mentioned in [31]. The isolation and characterization of Rhizobium strains from nodules of legumes grown in heavy-metal-contaminated soils were examined in many works, such as Sinorhizobium meliloti nodulating alfalfa [10] and Medicago lupilina [21], R. leguminosarum biovar trifolii isolated from clover nodules [1], and Agrobacterium tunefaciens found in the nodules of Lespedeza cuneata [23,32]. Nodulation tests suggested the existence of functional nodules in inoculated Sulla coronaria with rhizobial strains; it could be associated with symbiotic and nitrogen fixation abilities. In fact, nodulation of Sulla coronaria by Rhizobium sullae was demonstrated by [33] and [34] affirm that Gamma proteobacteria can nodulate the genus Hedysarum. The presence of many symbiotically effective Rhizobium in highly polluted soils seems to be very important [10]. In this work, four bacterial strains belonging to pseudomonas and Rhizobium sullae were selected and characterized in order to use them as symbiotic inoculums for Sulla coronaria, ameliorating the potential of phytostabilization of the plant in contaminated soils. The application of bacterial a consortium containing Pseudomonas as an endophytic bacteria and Rhizobium to improve the productivity of legumes was tested by [35], and its efficiency in the phytoremediation process had been previously demonstrated in many other works [11,27,36].
4.3 Accumulation of cadmium in cells of selected bacteria
The accumulation of cadmium in the whole cell was highest in the most Cd-resistant bacteria I10 (Pseudomonas sp.), suggesting the possible relationship with a mechanism of tolerance to heavy metals [23]. The intracellular Cd accumulation was greater than in the cell wall and it was enhanced with the time of incubation in the four strains; these phenomena demonstrated that Cd accumulation was related to the cell growth or to the increase in the biomass [21]. Moreover, other mechanisms of heavy metal tolerance were discovered such as biosorption and bioaccumulation both in internal and external spaces; these processes influence the bioavailability of heavy metal to plant [26].
4.4 Production of plant-growth-promoting substances by selected bacteria
In this work, the screened soil bacteria had the ability to produce IAA essentially in Pseudomonas sp. and Rhizobium sullae more than those reported in other heavy-metal-resistant bacteria [1,25]. The main effect of bacterial IAA is the enhancement of mineral and nutrient uptake, inducing bacterial proliferation on the roots [12].
In addition, these soil bacteria produced siderophores that indirectly affect the growth of plants; generally siderophores form a stable complex with iron and other heavy metals, resulting in an increase in metal concentration [26]. This phytohormone played an important role in plant–bacteria interactions and plant growth in metal contaminated soils [37]. Pseudomonas bacteria, especially P. fluorescens and P. Putida, are the most important kind of PGPR, which produce auxin and promote the yield [38]. The consortium of selected soil bacteria can be designed as a Cd-resistant PGPR that have the exceptional ability to ameliorate the growth of host plant in contaminated soils by various mechanisms such as nitrogen fixation, solubilisation of minerals, production of phytohormones and siderophores [37], as confirmed for Vicia faba by dual inoculation with rhizobium and PGPRs under copper stress [39].
4.5 Detection of Cd-resistance genes in the screened bacteria
PCR amplification and electrophoresis of some Cd-resistant genes selected showed a positive amplification of the Cd resistance gene with one or more primers in the four selected soil bacteria, suggesting that some primers were inappropriate. The sequence comparison showed a similarity with Cd-resistant genes of other bacteria, indicating that these genes were acquired in the selected soil bacteria by lateral gene transfer from other soil bacteria as has been proposed for some symbiotic genes [40,41]. These genes encoded certain enzymes of the mechanism of tolerance to Cd in bacteria.
5 Conclusion
This study suggested that the selected PGPRs, including Pseudomonas and Rhizobium sullae, were efficient and heavy metal tolerant; they accumulate Cd essentially in the intracellular compartment, produced plant-growth-promoting substances and revealed the presence of some genes responsible for the resistance to Cd. These characteristics suggest that this consortium could be a useful inoculum for Sulla coronaria to phytostabilize Cd-contaminated soils, and further studies are needed to clarify the mechanism. The prospective symbiotic system should be tested in contaminated areas.
Acknowledgement
This study was supported by the laboratory project entitled “Tolerance of Legumes to Biotic and Abiotic Stresses” financed by the Center of Biotechnology of BorjCedria.


