1 Introduction
Describing species is important to advancing biodiversity research at the local, regional, and broader scales, because species usually are the fundamental units in community ecology, biogeography, and conservation. To describe species, taxonomists typically use morphology as primary evidence and, increasingly often, genetic information to support morphology [1]. Although genetics have proven essential to delineate species in many instances, genetic information such as allele frequencies or DNA sequences is still exceptionally used as primary evidence in new species descriptions, in part if not mostly because of considerable peer pressure against DNA-based descriptions [1]. However, there is no objective reason to dismiss genetics as the primary source of characters for species delineation, description, and identification [1–3].
The blue-spotted maskray, previously N. kuhlii (Müller and Henle, 1841), has a wide Indo-West Pacific distribution, from eastern Africa to Japan [4–7]. It was hypothesized to be a species complex after a barcoding survey revealed several deeply divergent mitochondrial lineages [8]. The hypothesis of a species complex was subsequently validated [6,9]. This species complex consists of up to eleven parapatrically-distributed lineages representing separate species [6,9–12] of which nine have already been formally described [7,12–14]. These are N. australiae Last, White and Séret, 2016, N. bobwardi Borsa, Arlyza, Hoareau and Shen, 2017, N. caeruleopunctata Last, White and Séret, 2016, N. malaccensis Borsa, Arlyza, Hoareau and Shen, 2017, N. moluccensis Borsa, Arlyza, Hoareau and Shen, 2017, N. orientale Last, White and Séret, 2016, N. vali Borsa, 2017, N. varidens (Garman, 1885), and N. westpapuensis Borsa, Arlyza, Hoareau and Shen, 2017. Two other species in the genus, the nominal N. kuhlii from Vanikoro and N. trigonoides possess distinctive spot patterns [3,15,16] that tell them apart from the blue-spotted maskray as it was originally described by J. Müller and F.G.J. Henle [4]. Based on the only available information on colour patterns, one cannot exclude that N. kuhlii as it has been redefined by Last and co-authors [14] and N. trigonoides are synonyms [16]. The genetically distinctive Indian Ocean maskray reported in the recent phylogeographic literature [6,7,10] remains undescribed.
Some authors have attempted to use morphological characters as primary information for the description of cryptic species in the blue-spotted maskray complex [14], and a recent revision has ostensibly ignored genetic evidence (e.g., [17]) even though not a single morphological character among those utilized for the description or redescription of four species in the complex [14] was indisputably diagnostic of any of the species [7]. Also, the apportion of environmental vs. genetic determination in these morphological characters had not been evaluated [7]. Morphological diagnoses provided so far for the blue-spotted maskray [14] are therefore hardly relevant, as previously mentioned for the Himantura uarnak complex, another species complex among stingrays [3]. In contrast, DNA sequences offer a profusion of diagnostic characters in stingrays [3,6,9,14,18]. Species-specific haplogroups were observed in the blue-spotted maskray, based on DNA sequences [3,9].
The objective of the present paper is to formally describe the Indian Ocean maskray, primarily based on the mitochondrial DNA sequences of fresh specimens from the eastern coast of India and of previously-reported material from India and Tanzania.
2 Materials and methods
2.1 Material examined
Eleven specimens of the new species that were examined for spot patterns are listed in Table 1 (their pictures are provided in Supplementary Fig. S1). Specimen NKGMF-3 from the Gulf of Mannar, January 2017, to be designated as the holotype of the new species, was deposited at the Marine Biology Regional Centre (MBRC) in Chennai, India. The MBRC is part of the network of museums managed by, and an official repository of the Zoological Survey of India. Specimen NKGMF-1 from the Gulf of Mannar, January 2017, to be chosen as a paratype of the new species, was deposited at the Fish Genetics and Biotechnology (FGB) laboratory, ICAR-Central Institute of Fisheries Education (CIFE) in Mumbai, India. Specimens NITT1, NITT2 and NITT3 were collected in Tharivaikulum, Tuticorin, India, December 2017; all three specimens are preserved in formalin at the FGB laboratory ICAR-CIFE in Mumbai. Specimen IRD-20170720-A, purchased fresh from fishermen at the Malindi fish landing place in Zanzibar, July 2017, was preserved salted; it is currently stored at the IRD-La Valette Zoothèque in Montpellier, France. Voucher material of the new species also includes a specimen collected by APK and colleagues in Visakhapatnam, Andhra Pradesh, Bay of Bengal on 15 August 2011. The specimen was discarded but a sub-sample of tissue was registered at the Central Institute of Fisheries Education, Mumbai under No. VIZNK-01. A photograph of this individual is available from the Barcoding of Life Data systems database (BOLD; http://www.barcodinglife.com/; [20]) under accession No. BOLD:ACB9305. Photographs of blue-spotted maskrays from India deposited in FishBase [21] were not retained for the analysis of spot patterns because of insufficient resolution. Comparative material for the characterization of spot patterns consisted of specimens from N. australiae (N = 2, including holotype), N. bobwardi (N = 5, including holotype), N. caeruleopunctata (N = 2, including a paratype), N. malaccensis (N = 1, holotype), N. moluccensis (N = 2, including holotype and single paratype), N. orientale (N = 8, including holotype), N. vali (N = 3, including holotype), N. varidens (N = 6), and N. westpapuensis (N = 1, holotype) (Table 1).
Spot patterns on the dorsal side of 41 blue-spotted maskray (Neotrygon spp.) specimens sorted by species. Specimens from Padang and Kota Kinabalu, two specimens from the Solomon Islands [12] as well as ASIZP0073604, ASIZP0073605 and ASIZP0078458 were identified to species based on their sampling location [9]; all other specimens were identified to species based on their CO1 gene sequence [9]. L, left pectoral fin; R, right pectoral fin. Occipital mark, diffuse dark blotch at rear end of neurocranium that was categorized as either absent (0), weak (1), or conspicuous (2).
| Species, Specimen No. (field No.) |
Locality of collection, country | N ocellated blue spots | N dark speckles (< 1% DW) L, R |
N dark spots (> 1% DW) L, R |
Occipital mark | ||
| 1. Small L, R |
2. Medium L, R |
3. Large L, R |
|||||
| N. australiae | |||||||
| CSIRO 7016-01a | Weipa, Australia | 14, 12 | 17, 20 | 3, 0 | 8, 9 | 0, 0 | 0 |
| MZB-20863 (ENTTJS2) | Tanjung Sulamo, Indonesia | 25, 25 | 28, 32 | 1, 0 | 13, 6 | 0, 0 | 0 |
| N. bobwardi | |||||||
| MZB-20843 (ME3)a | Meulaboh, Indonesia | 28, 29 | 13, 11 | 0, 0 | 16, 16 | 0, 0 | 0 |
| 20150524-D | Padang, Indonesia | 5, 11 | 4, 2 | 0, 0 | 6, 6 | 0, 0 | 0 |
| 20150524-E | Padang, Indonesia | 16, 11 | 9, 8 | 0, 0 | 8, 15 | 0, 0 | 2 |
| 20150524-F | Padang, Indonesia | 20, 36 | 14, 8 | 0, 0 | 3, 0 | 0, 0 | 1 |
| 20150524-11 | Padang, Indonesia | 8, 8 | 2, 1 | 0, 0 | 6, 13 | 0, 0 | 2 |
| N. caeruleopunctata | |||||||
| CSIRO H 7850-01b | Sadeng, Indonesia | 25, 22 | 4, 1 | 0, 0 | 17, 30 | 0, 0 | 1 |
| 20080131_BL (NK-K-BL) | Bali, Indonesia | 21, 24 | 6, 5 | 0, 0 | 43, 54 | 0, 0 | 1 |
| MZB-22131 (BAL-S) | Bali, Indonesia | 31, 22 | 5, 9 | 0, 1 | 0, 1 | 0, 0 | 1 |
| N. indica sp. nov. | |||||||
| ZSI/MBRC/F.1495 (NKGMF-3)a | Gulf of Mannar, India | 9, 10 | 0, 0 | 0, 0 | 19, 34 | 3, 2 | 2 |
| CIFEFGB/NKGM1 (NKGMF-1)b | Gulf of Mannar, India | 27, 26 | 0, 0 | 0, 0 | 37, 22 | 4, 1 | 2 |
| VIZNK-01 (NKVSKP-1) | Visakhapatnam, India | 3, 12 | 1, 9 | 0, 0 | 13, 10 | 0, 0 | 2 |
| NKTTK-1 | Tuticorin, India | 30, 27 | 3, 3 | 0, 0 | 90, 86 | 0, 1 | 2 |
| NKTTK-2 | Tuticorin, India | 20, 8 | 0, 0 | 0, 0 | 24, 32 | 0, 1 | 2 |
| NKGMM-2 | Gulf of Mannar, India | 24, 26 | 7, 4 | 0, 0 | 30, 33 | 4, 1 | 2 |
| NKGMM-4 | Gulf of Mannar, India | 14, 18 | 8, 4 | 0, 0 | 40, 38 | 5, 6 | 2 |
| NITT-1 | Tuticorin, India | 28, 28 | 20, 14 | 0, 0 | 68, 73 | 1, 1 | 2 |
| NITT-3 | Tuticorin, India | 10, 32 | 9, 3 | 0, 0 | 34, 35 | 1, 3 | 2 |
| ZAN 1 | Zanzibar, Tanzania | 44, 51 | 29, 21 | 2, 2 | 10, 15 | 0, 0 | 0 |
| IRD-20170720-A | Zanzibar, Tanzania | 32, 35 | 6, 6 | 0, 0 | 16, 21 | 1, 0 | 2 |
| N. malaccensis | |||||||
| MZB-20847 (MSKL3)a | Kuala Lama, Indonesia | 2, 4 | 14, 10 | 0, 1 | 5, 3 | 0, 0 | 0 |
| N. moluccensis | |||||||
| MZB-20866 (ARA1)a | Tual, Indonesia | 36, 30 | 9, 9 | 0, 0 | 53, 53 | 0, 1 | 0 |
| MZB-20864 (AM1)b | Ambon, Indonesia | 9, 2 | 7, 12 | 0, 0 | 5, 2 | 0, 0 | 1 |
| N. orientale | |||||||
| CSIRO H7858-01a | Muara Kintap, Indonesia | 20, 14 | 15, 17 | 1, 1 | 2, 1 | 0, 0 | 0 |
| MZB-20852 (PB2) | P. Pabelokan, Indonesia | 5, 9 | 8, 3 | 1, 0 | 21, 17 | 0, 0 | 0 |
| MZB-20851 (PR) | Pulau Pari, Indonesia | 22, 18 | 9, 6 | 2, 2 | 12, 8 | 0, 0 | 0 |
| MZB-20850 (PN5) | Pulau Peniki, Indonesia | 12, 22 | 12, 11 | 2, 0 | 2, 1 | 0, 0 | 1 |
| BO424 | Tanjung Manis, Malaysia | 35, 41 | 22, 6 | 0, 0 | 16, 13 | 0, 0 | 0 |
| WJC-5367 | Kota Kinabalu, Malaysia | 17, 19 | 10, 8 | 0, 1 | 8, 4 | 0, 0 | 0 |
| WJC-5368 | Kota Kinabalu, Malaysia | 19, 20 | 6, 12 | 0, 0 | 6, 5 | 0, 0 | 0 |
| WJC-5369 | Kota Kinabalu, Malaysia | 20, 23 | 14, 10 | 0, 0 | 3, 2 | 0, 0 | 0 |
| N. vali | |||||||
| CSIRO H7723-01a | Honiara, Solomon Is. | 2, 4 | 1, 1 | 0, 0 | 3, 6 | 0, 0 | 0 |
| Randall ([19]: 18)c | Solomon Is. | 11 | 4 | 0 | 6 | 0 | 0 |
| Rosensteinc | Mbike wreck, Solomon Is. | 21 | 6 | 0 | 7 | 0 | 0 |
| N. varidens | |||||||
| ASIZP0073604 | Taiwan | 0, 1 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1 |
| ASIZP0073605 | Taiwan | 3, 2 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1 |
| ASIZP0073535 | Taiwan | 3, 1 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1 |
| ASIZP0078458 | Taiwan | 7, 16 | 0, 0 | 0, 0 | 0, 0 | 0, 0 | 1 |
| ASIZP0805866 | Taiwan | 4, 3 | 5, 6 | 0, 0 | 2, 7 | 0, 0 | 1 |
| N. westpapuensis | |||||||
| MZB-20867 (BK5)a | Biak, West Papua | 41, 37 | 25, 13 | 0, 2 | 10, 7 | 0, 0 | 1 |
a Holotype.
b Paratype.
c Left side only.
Five specimens, including four from India and one from Tanzania were measured according to [14] using a ribbon meter, a ruler, and a vernier calliper. Three of these, including the holotype and the paratype of the new species were X-ray photographed. Pectoral fin rays were counted from the X-ray photographs. Specimens of the new species that were characterized by their nucleotide sequences at loci CO1 and cytochrome b are listed in Table 2 and include 13 specimens from India and 6 specimens from Tanzania. Comparative material for the analysis of nucleotide sequences included blue-spotted maskray specimens listed in [9] and an additional specimen from Taiwan deposited in the fish collections of Academia Sinica, Taipei, and registered under No. ASIZP0805866.
Neotrygon indica sp. nov. specimens examined for nucleotide variation at mitochondrial loci CO1 and cytochrome b.
| Specimen No. (field No.) | Locality of collection, country | GenBank accession No. | Source | |
| CO1 | Cytochrome b | |||
| NBFGR:CHN 156 | Chennai, India | HM467799 | – | Mined from GenBank |
| NBFGR:CHN:R24 | Chennai, India | KF899609 | – | Mined from GenBank |
| NBFGR:CHN:R25 | Chennai, India | KF899610 | – | Mined from GenBank |
| NBFGR:CHN:R165 | Chennai, India | KF899611 | – | Mined from GenBank |
| NBFGR:CHN:NK13 | Chennai, India | KF899612 | – | Mined from GenBank |
| NBFGR:CHN:199 | Chennai, India | KF899613 | – | Mined from GenBank |
| – | Tamil Nadu, India | KR003770 | – | Mined from GenBank |
| ZSI/MBRC/F.1495 (NKGMF-3)a | Gulf of Mannar, India | MG017600 | MG017605 | Present study |
| NKGMM-2 | Gulf of Mannar, India | MG017601 | MG017606 | Present study |
| NKVSKP-1 (= VIZNK-01) | Visakhapatnam, India | JX978329, MG017602 | MG017607 | Mined from BOLD, Present study |
| NKVSKP2 | Visakhapatnam, India | MG017603 | MG017608 | Present study |
| NKVSKP3 | Visakhapatnam, India | MG017604 | MG017609 | Present study |
| NKTTK1 | Tuticorin, India | – | MG017610 | Present study |
| ZANZ 1 | Zanzibar, Tanzania | JX263421 | KU497907 | [10] |
| 80611 | Tanga, Tanzania | KC249906 | – | [6] |
| KNS-TZN1-52 | Pemba Island, Tanzania | KU498035 | KU497908 | [9] |
| KNS-ZAN3-86 | Pemba Island | KU498036 | KU497909 | [9] |
| KNS-ZAN4-71 | Pemba Island | KU498037 | KU497910 | [9] |
| KNS-ZAN5-80 | Pemba Island | KU498038 | KU497911 | [9] |
a Holotype.
2.2 Spot pattern analysis
The diameter of ocellated blue spots on the dorsal side of the left and right pectoral fins, relative to disc width (DW), was measured from photographs ([7,12,14,15]; Figs. 1 and 2; Supplementary Fig. S1). Ocellated blue spots were qualified as “small” when their maximum diameter was = 2% DW, “medium” when = 4% DW, and “large” when > 4% DW [15]. Dark speckles (= 1% DW) and dark spots (> 1% DW) were also counted on the dorsal surface of the disc. The counts did not include those speckles and spots located within the dark band around eyes that forms the mask [15]. The presence or absence of a darker occipital mark at the rear end of the neurocranium was also checked and categorized as “absent”, “weak”, or “conspicuous”. Spot patterns were compared among individuals through correspondence analysis (CA) [22]. CA was run using the FactoMineR package [23] under R [24].
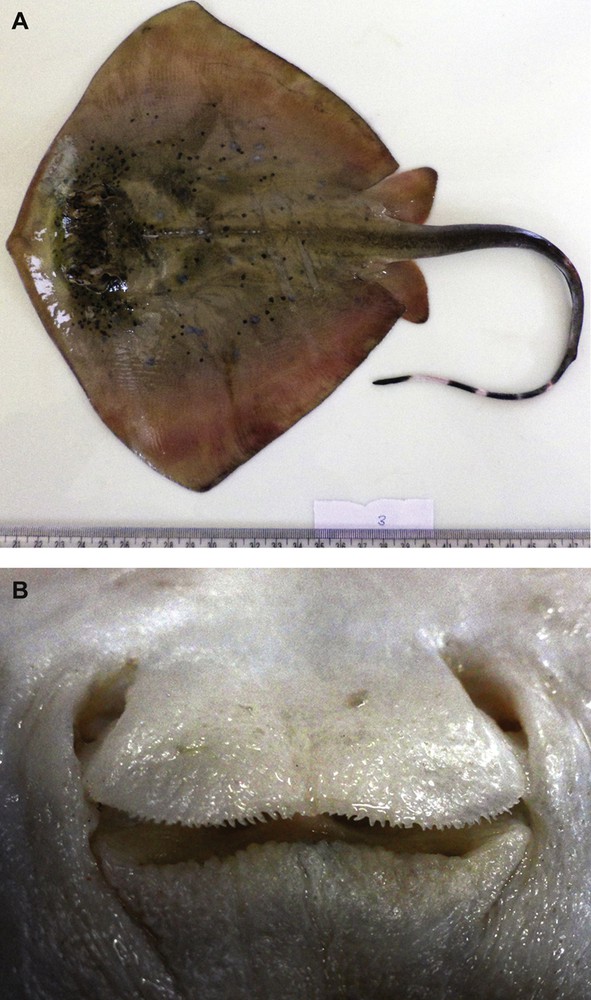
Holotype of Neotrygon indica sp. nov.: female specimen, 174 mm disc length from the Gulf of Mannar, Tamil Nadu, India (09.12°N 79.46°E) registered under No. ZSI/MBRC/F.1495 at the Marine Biology Regional Centre in Chennai, India. A. Dorsal side (photograph by RK). B. Oronasal region (photograph by APK).

Paratype of Neotrygon indica sp. nov.: female specimen No. CIFEFGB/NKGM1, 182 mm disc length from the Gulf of Mannar, Tamil Nadu, India (9.12°N 79.46°E) (photograph by RK).
2.3 Analysis of nucleotide sequences
The DNA of six specimens of the new species and of specimen ASIZP0805866 from Taiwan was extracted using the phenol-chloroform protocol and the DNEasy extraction kit (Qiagen GmbH, Hilden, Germany), respectively. A fragment of each the CO1 and the cytochrome b genes was amplified by polymerase chain reaction according to [9]. Nucleotide sequencing was done in both forward and reverse directions using the Sanger method. The consensus sequences were obtained after assembling the direct and reverse traces under BioEdit v. 7.1.11 [25]. Sequences were then edited and aligned under BioEdit. We chose as reference for numerating nucleotides the complete mitogenome sequence of Neotrygon orientale [26], accessible from GenBank (http://www.ncbi.nlm.nih.gov) under No. KR019777. Five out of 11 N. indica sp. nov. individuals characterized by their spot patterns (Table 1) had their nucleotide sequences at loci CO1 and/or cytochrome b examined (Table 2).
The phylogeny of concatenated CO1 + cytochrome b gene haplotypes was inferred using the maximum-likelihood (ML) method under Mega6 [27]. The most likely nucleotide-substitution model, which was determined according to the Bayesian information criterion, was the Tamura–Nei model [28] (TN93) where a discrete Gamma distribution (G = 0.76) was used to model evolutionary rate differences among nucleotide sites and invariable sites were allowed. The ML tree was rooted by choosing New Caledonian maskray N. trigonoides as outgroup [15]. The robustness of nodes in the tree was tested by bootstrap resampling.
Based on the larger number of CO1 gene sequences available ([9]; Table 2), a parsimony network was constructed to further explore the relationships among haplotypes of the new species, and with those of the geographically adjacent species N. bobwardi, N. caeruleopunctata and N. malaccensis. Median-joining parsimony analysis was done using Network [29] on the nucleotide sequence matrix of 57-individual CO1 gene haplotypes compiled from the present work and from previous reports [9], using the default settings of the program. This matrix comprised sequences from blue-spotted maskrays from India (N = 11) and Tanzania (N = 5) as well as from the geographically adjacent N. bobwardi from the Andaman Sea and Western Sumatera (N = 13), N. caeruleopunctata from southern Java and southern Bali (N = 18), and N. malaccensis from the Malacca Strait and the eastern Andaman Sea (N = 10). Prior to median-joining analysis, the sequence matrix was trimmed to a core length of 638 bp, between nucleotide sites 68 and 705 of the CO1 gene. The placement of the root was inferred from the maximum-likelihood tree of concatenated CO1 + cytochrome b gene sequences. Parsimony network analysis was similarly run on a matrix of 48 cytochrome b gene sequences, trimmed to a core length of 795 bp, between nucleotide sites 238 and 1032 of the cytochrome b gene. This comprised six sequences from India and five sequences from Tanzania (Table 2), as well as seven N. bobwardi, 17 N. caeruleopunctata, and 13 N. malaccensis.
The mean nucleotide distances within and between species were estimated from the same 57-individual CO1 and 48-individual cytochrome b sequence datasets using Mega6. According to the Bayesian information criterion, the best substitution model for the CO1 gene sequence dataset was K2 + G (G = 0.07). The best substitution model for the cytochrome b sequence data was HKY + G (G = 0.05), but the model used for the calculations was the second-best (TN93 + G), which, unlike the former, is available in Mega6.
2.4 Barcode index number assignment
The BOLD data systems distinguish clusters of sequences that qualify as operational taxonomic units, i.e. putative species using the Refined Single Linkage algorithm [30]. The latter “clusters sequences with high similarity and connectivity and separates those with lower similarity and sparse connectivity.” Each putative species thus flagged is allocated a unique barcode index number (BIN) in BOLD. Borsa et al. [7] have established the homology of BIN numbers with cryptic blue-spotted maskray lineages by visual inspection of the placement of the CO1 gene sequences retrieved from BOLD in a reference maximum-likelihood tree rooted by N. trigonoides.
2.5 Notice
The present article in portable document (.pdf) format is a published work in the sense of the International Code of Zoological Nomenclature [31] or Code and hence the new names contained herein are effectively published under the Code. This published work and the nomenclatural acts it contains have been registered in ZooBank (http://zoobank.org/), the online registration system for the International Commission on Zoological Nomenclature. The ZooBank life science identifier (LSID) for this publication is urn:lsid:zoobank.org:pub:10551DF9-4B93-40ED-8910-012C5DCF8B96. The information associated with this LSID can be viewed through any standard Internet browser by appending the LSID to the prefix “http://zoobank.org/”. The online version of the present paper is archived and available from the Comptes rendus Biologies (http://www.sciencedirect.com/science/journal/aip/16310691), bioRxiv, Cold Spring Harbor NY, USA (http://www.biorxiv.org/) and haL-IRD, France (http://www.hal.ird.fr/) repository websites.
3 Results and discussion
3.1 Spot patterns
Spot patterns of 11 N. indica sp. nov. specimens were summarized and compared to nine other species of the blue-spotted maskray complex (Table 1). N. indica sp. nov. specimens from India were characterized by a moderately large number of small ocellated blue spots (n = 15–57), a generally low number of medium-sized ocellated blue spots (n = 0–34), a total absence of large ocellated blue spots, a high number of dark speckles (n = 23–176), generally a few dark spots (n = 0–11; only exceptionally present in the other species examined), and a conspicuous occipital mark. These features were visible on the holotype (Fig. 1) and the paratype (Fig. 2) of the new species. One of the two individuals from Tanzania had similar features, but not the other (Table 1). Incidentally, all five N. varidens specimens examined here possessed at least one to several (n = 1–23) small ocellated blue spots, at variance with Garman's [13] original description, which states: “The species resembles D. kuhlii, but has […] no spots.” As the specimens examined here originated from Taiwan and not from Hong Kong, the type locality of N. varidens, it is possible that geographic variation in spot patterns exists within N. varidens. Four out of five N. varidens specimens remained distinct from all the other blue-spotted maskrays by the absence of dark speckles and spots (Table 1). N. varidens was also distinct from all its congeners except N. indica sp. nov. by the frequent absence of medium-sized ocellated blue spots (Table 1).
Correspondence analysis run on the whole matrix of individuals × characters (Table 1) plotted N. indica sp. nov. specimens as a cluster mostly distinct from the other species previously under N. kuhlii, albeit with some overlap (not shown). A second run focused on the specimens from the Indian Ocean, including all nine N. indica sp. nov. specimens from India, the two specimens from Zanzibar, and all available specimens of N. bobwardi, N. caeruleopunctata and N. malaccensis. Again, N. indica sp. nov. formed a cluster mostly distinct from the other species (Fig. 3; Supplementary Fig. S2). N. indica sp. nov. specimens tended to have a higher count of dark spots and dark speckles than the other three species (Fig. 3).
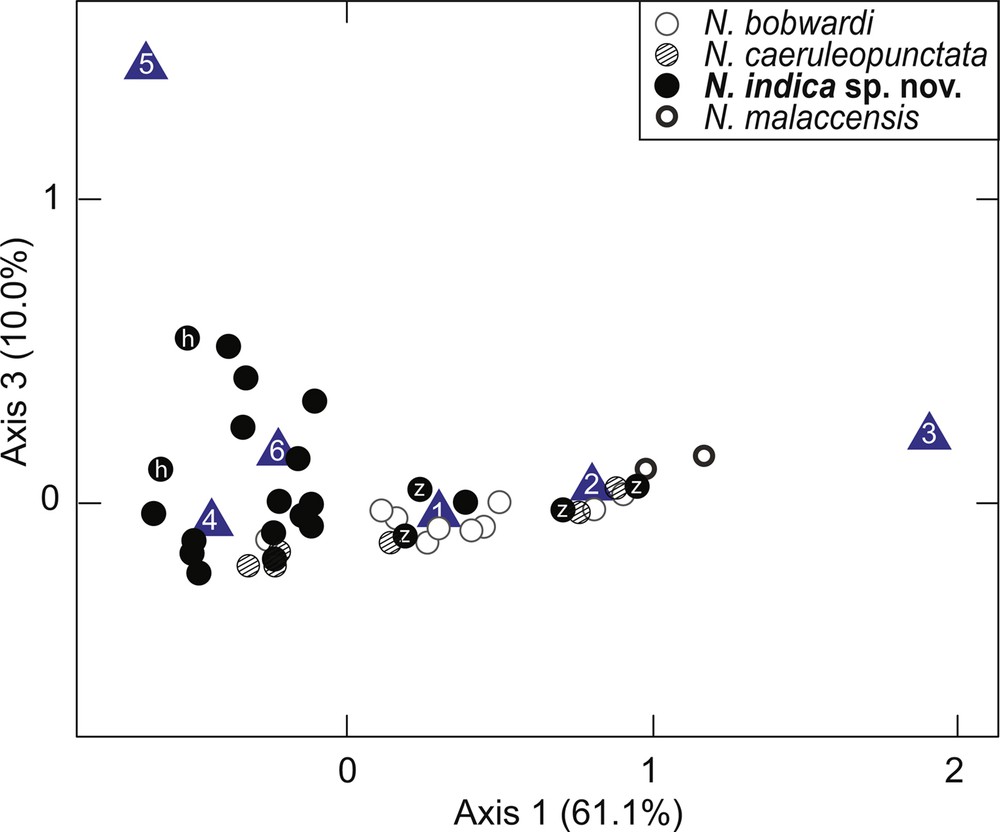
Correspondence analysis: projection of individuals characterized by counts of ocellated blue spots and dark spots and speckles, and by intensity of occipital mark on the dorsal side of a half-disc. Blue triangles indicate the position of the variables used to characterize spot patterns: 1. Small ocellated blue spots; 2. Medium-sized ocellated blue spots; 3. Large ocellated blue spots; 4. Dark speckles; 5. Dark spots; 6. Occipital mark. h Holotype; z sample from Zanzibar.
As shown by our results, some overlap in spot patterns was observed between some pairs of species. Also, one expects that with increased geographic sampling, based on the current sample sizes, more variation will show up within a species, hence further blurring the distinction between species and further relativizing diagnoses based on spot patterns.
3.2 Genetic distinctness of the Indian Ocean blue-spotted maskray
It has been recently reported that blue-spotted maskray populations from the Indian Ocean west of Bali (including N. bobwardi and N. indica sp. nov.) were “unresolved”, but hypothesized that they were closely related to N. caeruleopunctata [14]. Also, a single same BIN number (BOLD: AAA5611) has so far been allocated by BOLD to N. caeruleopunctata, N. indica sp. nov., and N. malaccensis [7], suggesting that the level of genetic differentiation at the CO1 locus is not sufficiently clear-cut to raise the three species, together with the closely related N. bobwardi to the level of fully distinct operational taxonomic units in the sense usually understood by the barcoding community. More in-depth investigation of the genetic relationships among populations within this group of geographically adjacent species from the Indian Ocean was warranted. Here, N. indica sp. nov. haplotypes formed a distinct haplogroup on the maximum-likelihood phylogenetic tree of concatenated CO1 + cytochrome b gene sequences (Fig. 4), confirming previous results [9]. A close-up within this haplogroup (Supplementary Fig. S3) revealed possible within-species phylogenetic structure, as it cannot be excluded that the Indian Ocean maskray comprises at least three sub-clades, including two from India and one from Tanzania. Testing this hypothesis would require more comprehensive and denser sampling of the wide Indian Ocean. Future, additional sampling effort is needed to investigate the phylogeographic structure of the Indian Ocean maskray in more depth.

Blue-spotted maskray Neotrygon spp. Simplified maximum-likelihood tree of concatenated CO1 + cytochrome b gene fragments, rooted by N. trigonoides [15] showing the placement of the N. indica sp. nov. lineage. Terminal branches were collapsed into triangles whose width is proportional to the number of individuals sequenced and whose depth represents the genetic variability within a species. Numbers at a node are bootstrap scores, from 600 bootstrap resampling runs under Mega6 [27]. For the same tree with all terminal branches apparent, see Supplementary Table S3.
Maximum-likelihood trees based on nucleotide sequences at the CO1 locus or the cytochrome b locus grouped N. indica sp. nov. haplotypes into a monophyletic lineage (Supplementary Fig. S4) or two poorly resolved haplogroups, where the haplotypes sampled in Visakhapatnam formed a distinct sub-clade (Supplementary Fig. S5). These minor differences in the topologies of the two trees presumably arose because of insufficient phylogenetic information borne by either gene fragment when considered separately. Nevertheless, the CO1 and cytochrome b median-joining parsimony networks (Fig. 5) both distinguished N. indica sp. nov. as a haplogroup separate from the other, geographically adjacent species. It is worth mentioning that in both CO1- and cytochrome b gene networks, haplotypes from Tanzania formed a distinct haplogroup within N. indica sp. nov. (Fig. 5).
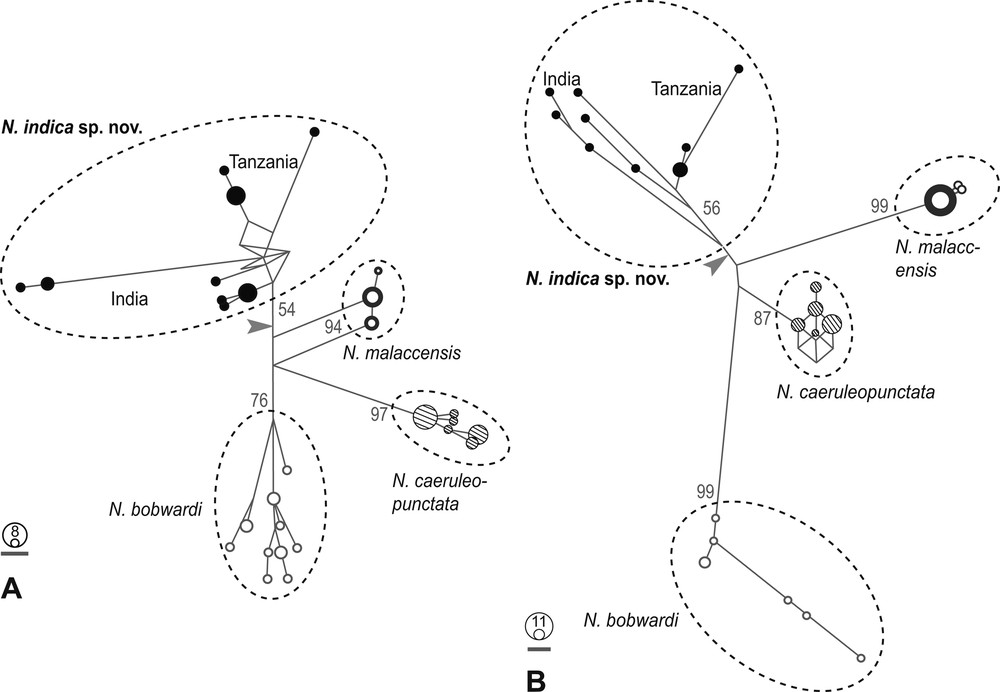
Genetic relationships of Neotrygon indica sp. nov. relative to other blue-spotted maskray Neotrygon spp. from adjacent regions in the Indian Ocean, based on mitochondrial haplotypes. The symbols for species are the same as those of Fig. 2. A. Median-joining parsimony network [29] of CO1 haplotypes (638-bp fragment of the CO1 gene). Groups of haplotypes delineated according to genetic proximity, with indication of the area of occurrence for N. indica sp. nov. The branch length is proportional to the number of mutational steps; the circles represent individual haplotypes, their area being proportional to their frequency in the total sample. The arrow indicates the possible placement of root, as inferred from the maximum-likelihood tree of concatenated CO1 and cytochrome b gene sequences (Fig. 4); numbers at nodes are bootstrap scores, obtained from the CO1 maximum-likelihood phylogeny (Supplementary Fig. S4). Scale bar: 1 mutational step. B. Homologous median-joining parsimony network based on a 795-bp fragment of the cytochrome b gene. Bootstrap scores from Supplementary Fig. S5.
Estimates of genetic distance between pairs of species in the Indian Ocean blue-spotted maskray group ranged from 0.020 to 0.025 at the CO1 locus and from 0.020 to 0.033 at the cytochrome b locus (Table 3). These values were all well above the highest estimates of genetic distance within a species (respectively, 0.012 and 0.014; Table 3). Therefore, the four species, albeit closely related, remained distinct from one another using either the standard CO1 barcode or the cytochrome b marker.
Blue-spotted maskray Neotrygon spp. from the Indian Ocean and geographically adjacent Andaman Sea. Estimates (mean ± SD) of nucleotide distances between pairs of, and within blue-spotted maskray species at loci CO1 and cytochrome b.
| Marker, Species |
Within-species | Species | ||
| N. indica | N. bobwardi | N. caeruleopunctata | ||
| CO1 | ||||
| N. indica | 0.012 ± 0.003 | – | ||
| N. bobwardi | 0.009 ± 0.003 | 0.025 ± 0.006 | – | |
| N. caeruleopunctata | 0.002 ± 0.001 | 0.022 ± 0.007 | 0.024 ± 0.007 | – |
| N. malaccensis | 0.001 ± 0.001 | 0.020 ± 0.006 | 0.020 ± 0.006 | 0.022 ± 0.008 |
| Cytochrome b | ||||
| N. indica | 0.014 ± 0.004 | – | ||
| N. bobwardi | 0.006 ± 0.002 | 0.033 ± 0.007 | – | |
| N. caeruleopunctata | 0.002 ± 0.001 | 0.020 ± 0.005 | 0.026 ± 0.006 | – |
| N. malaccensis | 0.000 ± 0.000 | 0.026 ± 0.006 | 0.026 ± 0.009 | 0.021 ± 0.007 |
So far, only two nuclear markers have been made available for maskrays Neotrygon spp. [6,9], and neither is variable enough to distinguish cryptic Neotrygon spp. as reciprocally monophyletic lineages. From the nuclear results synthesized by [9], it was nevertheless possible to distinguish Neotrygon indica sp. nov. from N. australiae, N. leylandi, N. moluccensis, N. orientale, N. picta, N. trigonoides, N. varidens and N. westpapuensis, but not from N. caeruleopunctata, and N. malaccensis.
3.3 Mitochondrial DNA sequences as reliable taxonomic characters
Here, useful characters to distinguish N. indica sp. nov. from its blue-spotted maskray congeners derived from, in part (1) the analysis of spot patterns and, more accurately (2) molecular genetics. Although spot patterns proved somewhat helpful to tentatively characterize species including N. indica sp. nov., they were not fully diagnostic. Therefore, we chose to primarily base the description and diagnosis of the new species on the more powerful nucleotide sequence of its mitochondrial DNA, as done previously with other species of the genus Neotrygon [7,12,15]. We have shown that meristics and morphometrics the way they were used in taxonomic descriptions of cryptic species in the blue-spotted maskray complex [14] hardly provided any diagnostic character [7]. Part of the problem may stem from the fact that so far individual values of meristic and morphometric parameters have not been made available to the community except for a few type specimens [14]. Values averaged over a number of individuals [14] suppress information on interindividual variation and are of limited interest in multivariate meristics and morphometrics. Here, we present morphometric values for four specimens from Tuticorin, India, and one specimen from Zanzibar, Tanzania (Table 4), as a contribution to a future morphometric database of Neotrygon spp. specimens. We also present counts of pectoral fin rays for three specimens from Tuticorin (see Supplementary Figs. S6–S8). It is hoped that in the future the database will include a sufficiently high number of specimens from this and other species in the genus Neotrygon so as to enable, if needed, meaningful morphometric analyses.
Morphometric measurements [14] for four Neotrygon indica sp. nov. specimens from the eastern coast of India (holotype, paratype, NITT1, NITT2) and one specimen from Tanzania (IRD-20170720-A). All measurements, except disc width and number of caudal stings expressed as percentage of disc width.
| Parameter | Specimen | ||||
| Holotype | Paratype | NITT1 | NITT2 | IRD-20170720-A | |
| Disc width (mm) | 236 | 213 | 314 | 307 | 300 |
| Total length | 169.5 | 183.1 | 191.1 | 198.7 | – |
| Disc length | 80.5 | 84.5 | 79.6 | 81.4 | 80.4 |
| Distance from tip of snout to pectoral fin insertion | 69.9 | 70.4 | 73.9 | 76.5 | 71.9 |
| Distance from tip of snout to eye | 15.3 | 16.0 | 15.9 | 16.3 | 13.8 |
| Mouth width | 10.6 | 8.5 | 8.3 | 9.1 | 10.0 |
| Nostril length | 6.8 | 4.7 | 5.1 | 5.9 | 4.2 |
| Distance between nostrils | 11.9 | 14.1 | 9.6 | 10.7 | 7.5 |
| Head length | 40.3 | 39.9 | 39.8 | 42.3 | 40.8 |
| Eye length | 6.4 | 6.6 | 6.4 | 6.5 | 5.4 |
| Orbit diameter | 10.2 | 9.4 | 7.6 | 7.8 | 7.1 |
| Inter orbital width | 7.6 | 7.0 | 7.3 | 7.2 | 7.1 |
| Inter ocular width | 16.9 | 18.8 | 12.7 | 13.0 | 16.1 |
| Spiracle length | 7.6 | 8.0 | 7.6 | 7.2 | 7.1 |
| Interspiracular width | 14.8 | 15.0 | 12.7 | 14.3 | 15.6 |
| Orbit and spiracle length | 9.3 | 14.1 | 19.7 | 13.0 | 10.0 |
| Orbit to pectoral fin insertion | 55.9 | 46.9 | 51.0 | 53.7 | 52.2 |
| Nasal curtain length | 5.9 | 4.7 | 6.4 | 6.5 | 4.6 |
| Nasal curtain width | 9.3 | 8.5 | 8.9 | 9.1 | 9.4 |
| Width of 1st gill slit | 4.2 | 3.3 | 3.2 | 3.3 | 3.3 |
| Width of 3rd gill slit | 3.0 | 3.3 | 3.2 | 3.3 | 3.8 |
| Width of 5th gill slit | 1.7 | 2.3 | 2.2 | 2.3 | 2.1 |
| Distance between 1st and 5th gill slits (left) | 13.6 | 14.1 | 19.1 | 19.5 | 14.2 |
| Distance between 1st and 5th gill slits (right) | 13.6 | 14.1 | 13.4 | 13.0 | 14.2 |
| Distance between 1st gill slits | 16.9 | 16.9 | 14.9 | 16.2 | 17.5 |
| Distance between 5th gill slits | 11.0 | 9.5 | 8.3 | 8.4 | 9.6 |
| Pre-nasal length of snout | 9.3 | 5.7 | 9.6 | 9.8 | 5.6 |
| Distance from tip of snout to cloaca origin | 62.7 | 67.6 | 70.1 | 71.7 | 71.8 |
| Distance from pectoral fin insertion to base of 1st sting | 44.5 | 37.6 | 38.2 | 35.8 | – |
| Distance from origin of cloaca to base of 1st sting | 42.4 | 44.6 | 43.0 | 42.3 | – |
| Cloaca length | 7.2 | 6.6 | 7.3 | 7.5 | 4.6 |
| Tail width at axil of pelvic fin | 8.5 | 9.4 | 8.3 | 9.8 | 9.4 |
| Tail width at base of 1st sting | 2.3 | 6.6 | 3.8 | 4.6 | – |
| Tail height at axil of pelvic fin | 5.5 | 6.6 | 4.8 | 4.9 | 4.6a |
| Tail height at base of 1st sting | 2.6 | 2.3 | 2.5 | 2.9 | – |
| Pelvic fin (embedded) length | 14.4 | 13.6 | 15.9 | 14.7 | 14.7 |
| Width across pelvic fin base | 19.9 | 18.8 | 19.1 | 17.9 | 22.1 |
| Greatest width across pelvic fin | 33.9 | 42.3 | 41.4 | 44.3 | 43.0 |
| Clasper length from pelvic axil | – | – | 15.6 | 16.6 | – |
| Post-cloacal clasper length | – | – | 20.4 | 22.8 | – |
| Tail width at base of sting | 3.4 | 3.8 | 7.6 | 7.8 | – |
| Tail height at base of sting | 2.5 | 2.8 | 3.8 | 3.9 | – |
| Disc thickness | 14.8 | 14.1 | 14.3 | 13.7 | 13.5a |
| Number of caudal stings | 1 | 1 | 2 | 2 | – |
| Length of 1st sting | – | 14.1 | 16.2 | 15.0 | – |
| Length of 2nd sting | – | – | 14.3 | 18.6 | – |
a Measured after the specimen had been salted.
4 Taxonomy
Neotrygon Castelnau, 1873 [32]. Characterized by a mask-shaped colour pattern around the eyes, a narrow internasal curtain, and a short tail that possesses well-developed dorsal and ventral skin folds [33]. Forms a distinct clade in the mitochondrial tree of Dasyatidae [6,18,26].
Blue-spotted maskray complex, Neotrygon spp., previously N. kuhlii. Characterized by polygonal shape and blue spots [4]. Blue-spotted maskray haplotypes cluster as a single clade in the mitochondrial phylogeny of the genus (figure 2b of [6,12]).
N. indica sp. nov. (Fig. 1A, B; Fig. 2; Supplementary Fig. S6); urn:lsid:zoobank.org:act:99F4F1A4-D5F6-4379-BC4E-7FB1E74CFA58. Trygon kuhlii [5]; presumably clade Neotrygon kuhlii 3 of Naylor et al. [18]; N. kuhlii [15]; N. kuhlii haplogroup I [10,11]; N. kuhlii clade 8 [6]; Clade I [9]; Indian Ocean maskray [7]. Also, BIN number BOLD:AAA5611 in BOLD.
4.1 Material examined
All specimens examined to characterize spot patterns are listed in Table 1. Specimens characterized morphometrically are listed in Table 4. The list of specimens characterized by their nucleotide sequence at mitochondrial loci CO1 and cytochrome b is provided as appendix A of [9]. Additional N. indica sp. nov. specimens sequenced at the CO1 and cytochrome b loci are listed in Table 2.
4.2 Types
A female specimen, 236 mm disc width (Fig. 1A) from the Gulf of Mannar, Tamil Nadu (9.12°N 79.46°E), collected at the Inico Nagar, Tuticorin fish landing centre by RK and Satish on 13 January 2017 is here designated as the holotype of Neotrygon indica sp. nov. The specimen was deposited by APK at the MBRC in Chennai, India on 12 May 2017. The specimen has registration No. ZSI/MBRC/F.1495.
A female specimen, 213 mm disc width (Fig. 2) fished from the Gulf of Mannar, Tamil Nadu (9.12°N 79.46°E) using bottom-set gillnets, collected by RK on 25 January 2017 is here designated as the single paratype of the new species. This specimen, labelled CIFEFGB/NKGM1 was subsequently deposited by APK at the Fish Genetics and Biotechnology laboratory, ICAR-Central Institute of Fisheries Education, Mumbai, India.
4.3 Description of holotype
The partial CO1 gene sequence of the holotype, comprised between nucleotide sites 51 and 722 of the gene, was 5'- C A C C C T T T A T T T A G T C T T T G G T G C A T G A G C A G G G A T A G T A G G C A C T G G C C T C A G T T T A C T T A T C C G A A C A G A A C T A A G C C A A C C A G G C G C T T T A C T G G G T G A T G A T C A A A T T T A T A A T G T A A T C G T C A C T G C C C A C G C C T T C G T A A T A A T C T T C T T T A T A G T A A T G C C A A T T A T A A T T G G T G G A T T T G G T A A C T G A C T A G T G C C C C T G A T A A T T G G G G C T C C G G A C A T A G C C T T T C C A C G A A T A A A C A A C A T A A G T T T T T G A C T T C T A C C T C C C T C A T T C C T A T T A C T G C T A G C C T C A G C A G G A G T A G A A G C C G G G G C C G G A A C A G G T T G A A C A G T T T A T C C C C C A T T A G C T G G T A A T C T A G C A C A T G C C G G A G C T T C T G T A G A C C T T A C A A T C T T C T C T C T T C A C C T A G C A G G T G T T T C C T C T A T T C T G G C A T C C A T C A A C T T T A T C A C A A C A A T T A T T A A T A T A A A A C C A C C T G C A A T C T C C C A G T A T C A A A C C C C A T T A T T C G T C T G A T C T A T T C T T G T T A C A A C T G T A C T T C T C C T G C T A T C C C T A C C A G T C T T A G C A G C T G G C A T T A C T A T A C T C C T C A C A G A C C G A A A T C T T A A T A C A A C T T T C T T T G A C C C A G C T G G G C G A G G A G A T C C C A T T C T T T A C C A A C A C C T C T T C T G A T T C T T T G G C C A -3'. This sequence has No. MG017600 in GenBank.
The partial cytochrome b gene sequence of the holotype, comprised between nucleotide sites 238 and 1032 of the gene, was 5'- A T C C G C A A T A T T C A C G C T A A C G G C G C C T C A A T A T T C T T C A T C T G T G C T T A T C T C C A T A T T G C T C G A G G A C T T T A C T A T G G C T C C T A C C T C A A T A A A G A A A C C T G A A A C A T C G G A G T A A T T A T C C T A G T G T T A C T A A T A G C C A C C G C A T T T G T A G G C T A T G T T C T C C C A T G A G G A C A A A T A T C A T T C T G A G G G G C A A C C G T T A T T A C C A A C T T G C T A T C A G C C C T C C C C T A T A T T G G A G A C A T G T T A G T T C A A T G A A T C T G A G G A G G C T T C T C A A T T G A C A A T G C A A C A C T A A C T C G A T T T T T C A C A T T T C A T T T T C T A T T T C C C T T T G T A A T T G C A G C T C T T A C T A T A A T T C A C C T T C T C T T C C T T C A T G A A A C A G G T T C T A A C A A C C C A A C C G G A C T C T C A T C C A A C A T A G A C A A A A T C C C G T T T C A T C C T T A T T A T A C A T A T A A A G A T C T A G T A G G C T T C T T C A T C C T T C T A A T A C T A C T A A C T C T A C T T G C C T T A T T T A C A C C A A A C C T C C T A G G A G A T A C A G A A A A C T T T A T C C C A G C C A A C C C C C T C G T C A C A C C T C C C C A T A T T A A A C C A G A G T G A T A C T T C T T A T T T G C C T A C G C T A T T C T A C G C T C C A T C C C C A A T A A A C T A G G A G G A G T C C T A G C C C T C G C C T T C T C A A T C T T T A T C C T G C T A C T A G T C C C C A T T C T T C A C A C C T C C A A A C A A C G A A G C C T C A C C T T C C G T C C A A T T A C A C A A C T C C T A T T C T G A C T C T T A G T G G C A A A T A C A A T C A T C C T A A C A T G A A T C G G C G G C C A A C C C G T A -3'. This sequence has No. MG017605 in GenBank.
Ocellated blue spot, dark spot, and dark speckle counts on the dorsal side of the holotype are presented in Table 1. The meristic details of the holotype, which were scored from X-ray photographs (Supplementary Fig. S6), were the following: number of pectoral fin radials 106–108, including propterygion 47–51, mesopterygion 12, and metapterygion 45–47. Morphometric measurements on the holotype are presented in Table 4.
4.4 Measurements on paratype
Similarly, spot pattern details on the dorsal side of the paratype are presented in Table 1 and its meristic details were scored from X-ray photographs (Supplementary Fig. S7). The number of pectoral fin radials was 108–110, including propterygion 45–47, mesopterygion 14–15, and metapterygion 47–50. Morphometric measurements are presented in Table 4.
4.5 Diagnosis
N. indica sp. nov. from India is differentiated from all species of the blue-spotted maskray described so far (N. australiae, N. bobwardi, N. caeruleopunctata, N. malaccensis, N. moluccensis, N. orientale, N. vali, N. varidens and N. westpapuensis) by a combination of low number of medium-sized ocellated blue spots, total absence of large ocellated blue spots, high number of dark speckles, frequent occurrence of a few dark spots, and conspicuous occipital mark. However, this diagnosis did not apply to the two specimens sampled from Zanzibar whose spot patterns were examined. N. indica sp. nov. is distinct from N. kuhlii and N. trigonoides by the absence of the pair of conspicuous scapular blotches characteristic of these two species [15,16], although specimens of N. indica sp. nov. often possess a pair of diffuse darker marks in the scapular region.
Fifteen out of 19 individuals of N. indica sp. nov. sequenced at the CO1 locus possessed T at nucleotide site 607 of the CO1 gene, a character that was otherwise present in only two out of 130 N. orientale individuals and absent in all other cryptic species of the blue-spotted maskray (supplementary table S1 of [7]; present study). N. indica sp. nov. was further distinguished from other species previously referred to as N. kuhlii by A at nucleotide site 70 of the cytochrome b gene (based on five individuals from Tanzania). Eight out of 11 individuals of N. indica sp. nov. also possessed T at nucleotide site 284 of the cytochrome b gene, a character that was otherwise present in only one out of 77 N. orientale individuals and absent in all other cryptic species of the blue-spotted maskray (supplementary table S2 of [7]; present study).
4.6 Distribution
The type locality of N. indica sp. nov. is the Gulf of Mannar in Tamil Nadu on the eastern coast of the Indian subcontinent. Based on the collection of voucher specimens from the present study, the distribution of N. indica sp. nov. includes the coasts of Andhra Pradesh and Tamil Nadu states of India, from approximately 17.7°N to 8.8°N. Based on the material genetically examined thus far, the distribution of N. indica sp. nov. also includes the Indian coast of the Laccadives Sea (Kerala) and Tanzania ([6,7,9,10]; present study).
4.7 Etymology
The epithet indica is the Latin feminine adjectival form of the name of the country of the type locality, India.
4.8 Proposed vernacular names
Indian Ocean blue-spotted maskray (English); Neeli Nishan Pakat (Hindi); Pulli Thirukhai (Tamil); “raie pastenague masquée à points bleus de l’océan Indien” (French).
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
We thank all the people involved in the collection of specimens, tissue samples and photographs of blue-spotted maskray reported prior to the present study [9,12,15,34]. We thank W.-J. Chen and F. Giancarlo for providing additional photographs from, respectively, Kota Kinabalu and Padang. We are grateful to Dr. Gopal Krishna, Dr. Aparna Chaudhari and Dr. P. Gireesh Babu from ICAR-CIFE Mumbai for support, to Mr. Anand Kumar, Mr. Surender, Mr. Rajendra Kumar from the Marine Biological Research Centre, ZSI, Chennai for help in taking X-ray images, to Mr. Daniel from TNFU, Chennai for help in collecting samples, to Mr. A. Khatib from the Fisheries Department of Zanzibar for assistance at the Malindi fish landing place, Zanzibar. Photographs of N. varidens were downloaded from the Academia Sinica Fish Database website (http://www.fishdb.sinica.edu.tw/). Pre-mid-twentieth century books and articles were consulted online from the Biodiversity Heritage Library website (http://www.biodiversitylibrary.org/). Funded by ICAR-CIFE (India), the Agricultural Technology Research Institute of Taiwan and IRD (France). Insightful comments were made by two reviewers. Designed the study: APK, PB; did the experiments: APK, PP, PB; analysed the data: APK, PB; contributed reagents/materials/analysis tools: APK, RK, KNS, PB; wrote the paper: PB. All co-authors read, edited, and approved the final manuscript.


