1 Introduction: symbiosis as a driving force of evolution
Symbiosis, as first coined by Anton de Bary in 1879 [1], is an association between two or more organisms belonging to different species. This initial definition, which has experienced a major shift towards the concept of mutualism during the 20th century, did not originally mention any attribute regarding the durability or the extended phenotype of the association. Nowadays, biologists are aware that the outcome of symbiotic associations is often context-dependent and that symbiosis encompasses a continuum of interactions ranging from mutualism to parasitism [2–8]. Symbiosis is widespread in nature and basically there is no “aseptic” organism on earth. It can be terrestrial (e.g., lichens, plant mycorrhiza and rhizobia, microbiota of vertebrates and invertebrates), pelagic (e.g., corals, mollusks) and in extreme aphotic environments (e.g., giant tube worms, shrimps). It occurs at different levels of host integration: extraorganismal (e.g., marine nematode ectosymbiosis), intraorganismal (e.g., gut microbiota endosymbiosis), intracellular (e.g., insect and plant endosymbiosis) and ultimately as an organelle in eukaryotic cells (e.g., mitochondria and plastids). Moreover, while the concept of association has long been interpreted as a system of mutual dependence, where each part devotes itself to a given function and yields benefits to the other parts, current knowledge emphasizes that animals and plants rely for their reproduction and development on a patchwork of microbial associates, the microbiome, with which they constitute a biological unit or holobiont, subject to natural selection [9,10].
Symbiosis is currently accepted as a driving force in Evolution, notably by creating life diversity, increasing species adaptation and generating biological novelty. In 1910, the Russian botanist Konstantin Mereschkowsky [11] articulated the term “symbiogenesis” to explain the origin of eukaryotic cells by merging of prokaryotes. In 1918, the French biologist Paul Portier [12] published the book Les Symbiotes, in which he claimed that mitochondria, the cell organelles that generate ATP by oxidative processes, have originated from a symbiosis process; but it is the Russian botanist Boris Kozo-Polyansky [13] who has first recognized that life complexity occurs through the union between organisms, and explained in 1924 the symbiogenesis theory in terms of Darwinian evolution. Symbiogenesis was, however, dismissed for about a half century until the 1960's, when the American microbiologist Lynn Margulis provided microbiological evidence, even though controversial in some aspects, to strengthen the symbiogenesis theory, which she renamed the “Serial Endosymbiotic Theory”. Thanks to the inputs of several authors, including J. Maynard Smith, M. McFall-Ngai, P. Bonfante-Fasolo, M. Beth Saffo, P. Nardon, J. Sapp, N. Moran, P. Baumman, S. Scannerini, W. Schwemmler, A. Moya, A. Douglas, T. Fukatsu and H. Ishikawa, symbiogenesis and the neo-Darwinian concept of evolution are now fully integrated, in that most of biologists support the idea that “Hereditary endosymbiosis, supplemented by the gradual accumulation of heritable mutation, results in the origin of new species and morphological novelty” [14].
2 Insect endosymbiosis as a case study
To tackle the cellular processes involved in symbiogenesis and identify mutational and gene rearrangement events that have shaped major biological innovations, one approach is to investigate morphological structures and more generally biological innovations that have emerged along host-symbiont interaction in different plant and animal clades, such as plant nodules, squid light organs, or insect bacteriomes. Insects are among the most successful groups on our planet. Emerged at the Devonian (around 400 Myr), they have succeeded in adapting to numerous ecological niches and environmental conditions. Today, they represent probably the most diverse animal group and display many adaptive traits, including their relative small size that allows them to colonize microhabitats, their flight ability that facilitates geographic spreading and predator escaping, their exoskeleton (the cuticle) that prevents desiccation and limits pathogen infection, the metamorphosis phenomenon that improves adaptation and decreases intraspecific competition, their high fecundity that increases species survival, and their ability to associate and coevolve with flower plants. Remarkably, in addition to a few other invertebrates, insects are prone to house vertically transmitted intracellular microbial partners (endosymbionts) that provide them with nutrients lacking or limited in some habitats, improving thereby their fitness and invasive power [15]. To maintain permanently the endosymbionts and favor their maternal transmission through host generations, a compartmentalization strategy has been selected in most insects, consisting in the endosymbiont seclusion within specific cells, the bacteriocytes, which form the bacteriome organ in some insect species. This compartmentalization prevents the direct contact of endosymbionts with the host systemic immune system, while insect cellular and humoral immunity retains its ability to cope with infections by other bacterial intruders.
The team “Symbiosis and Immune Signaling” from INSA-Lyon conducts its research on insect bacteriomes. We aim to unravel the major “evolutionary strategies” that allow symbiont maintenance and transmission across host generations, to investigate whether symbiotic evolutionary constraints have shaped gene sequences and created new functions, and to elucidate how these emerging functions are involved in host homeostasis and endosymbiont control. The cereal weevil Sitophilus oryzae association with the endosymbiont Sodalis pierantonius displays many interesting features as a model to address these questions (Fig. 1). First, weevils house one endosymbiont only, constituting a simple one-to-one symbiont–host model system. The lab strain that we use is free of Wolbachia, a common intracellular bacterium found in arthropods and nematodes [16] and remarkably, no commensal bacteria have been identified in the gut of lab weevils, neither by PCR nor by cell imaging (unpublished data). Moreover, artificially non-symbiotic (aposymbiotic) viable weevils can be obtained [17], allowing for comparative studies between monosymbiotic and aposymbiotic insects within the same host species. Last, S. pierantonius has established symbiosis with weevil only recently: weevils from the Dryophthoridae family share a trophic endosymbiosis with diverse endosymbionts that have integrated symbiosis at different evolutionary periods [18–22]. The cereal weevils such as S. oryzae are unique as, contrary to their relatives, they would have established symbiosis recently (< 0.03 Myr) with S. pierantonius [23], whose genome, despite having lost many genes as compared to free-living relatives, has not experienced a size reduction as observed in other oldest endosymbiotic bacteria [24]. This evolutionary particularity of the S. oryzae/S. pierantonius association helps to decipher early mechanisms of endosymbiogenesis.

Endosymbiont and bacteriocyte dynamics during the cereal weevil life cycle. Fluorescent in situ hybridization microscopy showing Sodalis pierantonius endosymbiont (green). A: Permanent infection of oocytes (oo) by S. pierantonius (left panel; adapted from [57]); bacteriocyte differentiation in embryo (middle panel; adapted from [55]); bacteriome organ in larva (main panel). B: Young adulthood: S. pierantonius multiplies drastically in mesenteric bacteriomes. C: Adulthood: Gut-associated symbionts have been eliminated. L: larva; yA: young adult; 2-wA: 2-week old adult; oo: oocyte; b: bacteriocyte; Bact: bacteriome; ABact: adult bacteriome; fG: fore-gut; mG: midgut. Masquer
Endosymbiont and bacteriocyte dynamics during the cereal weevil life cycle. Fluorescent in situ hybridization microscopy showing Sodalis pierantonius endosymbiont (green). A: Permanent infection of oocytes (oo) by S. pierantonius (left panel; adapted from [57]); bacteriocyte differentiation in embryo ... Lire la suite
3 Immunity in an endosymbiotic system: a balancing act
The genome of S. pierantonius encodes secretion systems, regulatory elements and the enzymes involved in the synthesis of cell wall elements such as peptidoglycans and lipopolysaccharides, which are classical elicitors of innate immunity [24]. One essential question raised by this model is how the host immune responses are regulated to allow maintaining and controlling endosymbionts, while managing immune homeostasis and preventing a costly systemic activation of the immune response? Similar questions are raised with regards to gut bacteria commensalism and so far, most of the knowledge has come from this field of investigation [25–29]. In the fruit fly Drosophila melanogaster, which has emerged as a model to address these essential questions, two main responses control the gut bacterial load: the local production of reactive oxygen species [30,31], and the expression of antimicrobial peptides (AMPs), whose gene transcription is regulated by the Imd pathway [32,33]. The Imd pathway is activated through two main receptors, PGRP-LC and -LE [34,35], which detect the peptidoglycan of Gram-negative bacteria [36]. This recognition leads to the activation of a signaling cascade involving the intracellular protein IMD and leading to the activation and nuclear translocation of the NF-κB transcription factor Relish, which regulates the transcription of numerous genes, including AMP genes. The Imd pathway is tightly regulated at many transduction levels by various mechanisms, allowing a moderate response to commensal bacteria [37–44]. In contrast, only few data are available on immune regulations and intracellular symbiosis [45–52]. In this context, the cereal weevil is one of the most advanced models. In the last decade, we have shown that the weevil bacteriocyte immune response is modulated [53,54]. The bacteriome immune program consists in the strong expression of a unique AMP, Coleoptericin A (ColA), which controls endosymbiont growth and division, while the expression of other AMP genes, potentially harmful for the endosymbiont, remains low [54–57]. In vitro tests on ColA have demonstrated that a bactericidal activity against the Gram-negative bacteria Escherichia coli is only reached at relatively high concentrations of this peptide. At lower concentrations, ColA acts as bacteriostatic, and inhibits bacterial cell division, which generates the formation of filamentous gigantic bacteria, reminding the previously described shape of S. pierantonius [55]. The inhibition of ColA expression in the cereal weevil, using RNAi, decreased the endosymbiont size, strengthening the fact that ColA is involved in S. pierantonius gigantism in vivo. Most importantly, the inhibition of ColA expression also led to a loss of symbiotic control, with endosymbionts escaping the bacteriome and infecting the surrounding tissues. Therefore, ColA appears as an “endosymbiont's shepherd” and a key molecular player of their seclusion inside the bacteriome [55,57] (Fig. 2). Whether this function is the result of colA gene shaping during long-term host–symbiont coevolution is an intriguing hypothesis that is still under investigation. The data we have so far collected in favour of colA divergence rely on its comparison with colB, another coleoptericin gene present in the cereal weevil genomes and showing important sequence similarity to colA. However, while colA is highly expressed in the bacteriome, colB expression remains low in this organ in the absence of infection with free-living bacteria, similarly to the other AMPs studied. ColA and ColB also diverge in terms of biochemical activities: while both act as bacteriostatic or bactericidal depending on their concentration, ColB does not inhibit cell septation nor generate bacterial gigantism when incubated with E. coli, contrary to ColA. Further analysis of the two peptides’ bacterial interactors revealed several common bacterial substrates among E. coli proteins: OmpC, rp-L2 and EF-Ts. However, only ColA has evolved the specificity to interact with OmpA or GroEL. ColA-GroEL interaction was further confirmed on S. pierantonius chaperonin and ColA was shown to reach the bacterial cytosol where this interaction could occur [55,57]. Thus, these data suggest ColA as a good example of an evolutionary innovation driven by endosymbiogenesis, as they outline a model where colA sequence has evolved in favor of a “shepherd” function towards the endosymbiont, likely through the peptide ability to locate inside the bacterial cytosol, interact with the chaperonin GroEL, inhibit bacterial cytokinesis, and ultimately this results in bacteria seclusion inside the bacteriome, while maintaining a high endosymbiotic metabolic activity.
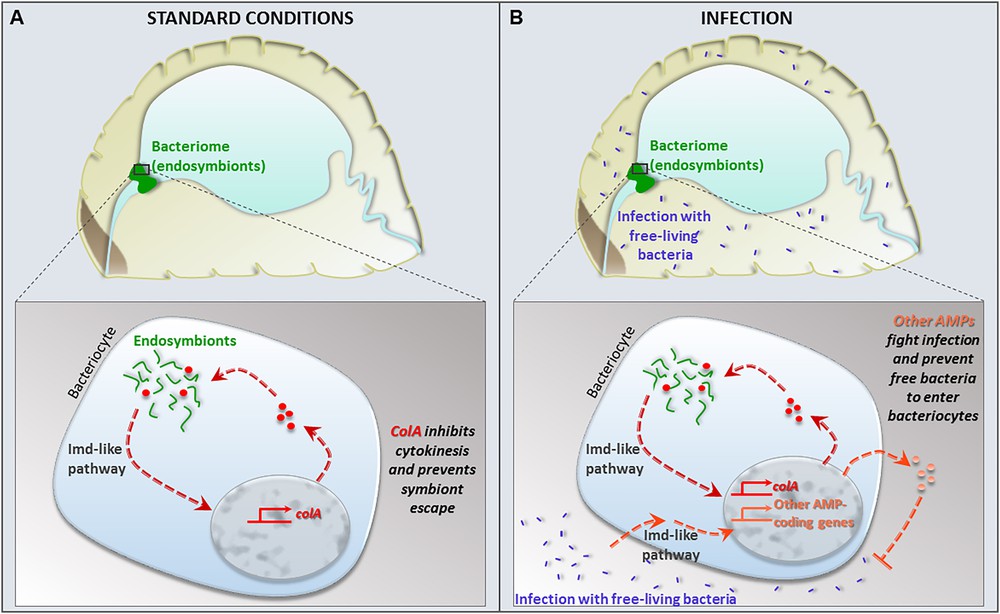
Model of endosymbiosis homeostasis under standard conditions and infection. A: under standard conditions, ColA AMP participates in the endosymbiont seclusion inside the bacteriocytes that form the bacteriome. colA is the only AMP-coding gene expressed under these conditions. B: upon infection with exogenous free-living bacteria, the bacteriome mounts an immune response that involves the induction of colA and other AMP-coding gene expression.
The precise mechanisms of bacteriocyte immunomodulation (i.e. high colA expression and low expression of other AMPs) are still under investigation. We have recently demonstrated that the weevil genome likely encodes all the genes required for a functional IMD-like response (Vargas et al., in prep.) and that the bacteriome organ expresses the imd gene [56]. Moreover, we have shown that the bacteriome is immunoreactive and is able to activate the expression of a cocktail of AMP genes when the insect faces a systemic infection with pathogenic bacteria [58]. Therefore, we know that the specific immune program of the bacteriome that favors symbiosis homeostasis under standard (non-infectious) conditions is not the result of a lack of immune competency of this organ [59]. The bacteriome immune program allows both keeping endosymbionts in and pathogenic infectious bacteria out (Fig. 2). Intriguingly, we recently showed that the bacteriome immune responses to both endosymbionts (i.e. colA specific expression) and pathogens are regulated by an IMD-like pathway, involving both IMD and Relish proteins [60]. Nonetheless, distinct transcription factor combinations and/or other regulatory factors, including non-coding RNAs and epigenetics marks could also be involved [61].
4 Endosymbiosis, metabolism and immunity: towards an integrated view
Another main question of the field, relevant both for gut commensal symbiosis and nutritional endosymbiosis, is how microbiota, metabolism and immunity are interconnected [27,31]. We have recently addressed this question by first demonstrating that the weevil endosymbiont load is dynamic and modulated according to the host needs at key stages of its development [62]. At the larval stage, the endosymbiont load increases slowly and the endosymbionts are contained in a unique bacteriome. In contrast, multiple bacteriomes are found at the apex of the mesenteric caeca in adults (Fig. 1). In the emerging adults, the endosymbiont population drastically increases (> 15-fold in five days). This bacterial load growth was shown to match an important metabolic need of the host, which builds its protective shell, the cuticle, at this developmental stage and requires an increased amount of tyrosine and phenylalanine amino acids to do so [62]. Strikingly, once the cuticle is achieved, the endosymbionts are quickly eliminated in one week (Fig. 1). We have shown that this elimination does not involve the activation of local nor systemic immune responses [62,63], but instead relies on the activation of two cellular mechanisms, apoptosis and autophagy. Massive autophagy suggests that the host recycles the endosymbiont components, while “clean” cell death of bacteriocytes by apoptosis allows the “shutdown” of these “metabolic factories” without triggering a costly inflammatory immune response. These two processes also present the particularity of being cell-autonomous, which provides tissue-specificity to endosymbiont elimination: while gut-associated endosymbionts are eliminated, ovarian bacteriomes persist in females, from which endosymbionts are transmitted to the next generation. The complete elimination and recycling of gut-associated endosymbionts at two weeks into adulthood, while weevils live up to six months under lab conditions [62], is reminiscent of a “mutualistic trade-off” between the endosymbiont contributions to the host metabolism and the cost associated with the endosymbiont maintenance inside a bacteriome. It remains to elucidate how these canonical cell mechanisms are integrated in symbiosis homeostasis and synchronized with the host physiological needs at a key stage of development. Interestingly, apoptosis, which seems to be shared by all multicellular eukaryotes, has been proposed to originate from the endosymbiotic genesis of mitochondria [64]. Comparative genomics further suggested that many genes involved in apoptosis have evolved from bacterial genes acquired through the domestication of the pro-mitochondrial endosymbiont [65]. As for autophagy, one of its main functions is the elimination of dysfunctional mitochondria [66], and it has also been proposed to be a biological innovation related to the integration of the pro-mitochondrial endosymbiont [67]. Thus, the use of autophagy and apoptosis in the modulation of endosymbiotic load not only constitutes an example of “evolutionary innovation by recycling”, where coevolution with the nutritional endosymbiont would be the driven force of innovation, but more, the processes of autophagy and apoptosis themselves could constitute evolutionary innovations driven by the pro-mitochondrial endosymbiogenesis.
Conclusion
Growing evidences indicate that the coevolution of host and bacterial symbionts has led to a reciprocal shaping of both innate and adaptive immunity and the microbiota [68–72]. In line with that and to parallel the theory best developed by Lynn Margulis that symbiosis is a source of evolutionary innovation, our data indicate that endosymbiosis is a source of immune innovation. Host–endosymbiont coevolution appears to have selected a powerful strategy to control, protect and modulate the endosymbionts, through the organogenesis of a tissue dedicated to endosymbiont housing: the bacteriome. Symbiont compartmentalization prevents the activation of the host systemic immune response in the absence of other infections and by isolating the symbionts, protects them whenever this immune response is triggered. We have identified the antimicrobial peptide ColA as a key factor of this compartmentalization strategy, with ColA acting as a “guard” that keep endosymbionts shut in the bacteriome. Last, the seclusion strategy allows the insect to shut down rapidly and in a safe way the bacteriocytes/“metabolic factories” whenever their benefit is over. We have shown that the rapid and tissue-specific adjustment of the symbiont load at a given stage of development involves two canonical cell processes, autophagy and apoptosis, whose function in the control of insect endosymbionts can be considered as another evolutionary innovation. Deciphering how these mechanisms are interconnected and integrated with the host metabolism may open up a new avenue for the understanding of holobiont function and evolution.
Funding
This work was supported by the French ANR-10 BSV7- 170101-545 03 (ImmunSymbArt) and ANR-13-BSV7-0016-01 (IMet–Sym), and by INRA and INSA Lyon.
Acknowledgments
We are grateful to the Symbiosis and Immune Signaling team, whose work contributed to the data presented here, and more largely to the BF2i (“Biologie fonctionnelle insectes et interactions”) lab, for its past and present stimulating collaborators. We would also like to acknowledge the symbiosis research community as a whole, for its readiness to fruitful scientific discussions.


