1 Introduction
The ATP-Binding Cassette (ABC) transporter superfamily comprises ATP-dependent transmembrane proteins that facilitate the bidirectional transport of substrates through the intracellular membranes of mitochondria, endoplasmic reticulum, Golgi apparatus, lysosomes, and peroxisomes [1–3]. These are classified as full-transporters and half-transporters and are subdivided into seven subfamilies (A–G) based on their sequence similarity and the organization of their domains. Full transporters comprise two homologous halves and are characterized by two membrane-spanning domains (MSDs) and two nucleotide-binding domains (NBDs), with an arrangement of MSD1–NBD1–MSD2–NBD2. Half transporters, such as ABCG2, contain only one MSD and one NBD, which are about half the size of a full transporter [4–6]. Additionally, these transmembrane proteins need to dimerize to form a functional membrane transporter [7,8]. While the ABCG transporter subfamily has been associated with the movement of substrates such as steroids and lipids, the human ABCG2 polypeptide has been involved in the transport of antivirals, antibiotics, and chemotherapeutic agents [7,9].
It has been shown that the ABCG2 polypeptide is localized in the plasma membrane and is highly expressed in synctiotrophoblasts, kidney, hepatocytes, blood brain barrier, breast tissue, prostate epithelium, and intestinal epithelial cells, where it could regulate the uptake and metabolism of xenobiotics [4,8,10]; it has been suggested that, in these tissues, ABCG2 expression plays the protective role of the blood–tissue barrier [9,11,12]. In addition, there are experimental data that have associated ABCG2 with the endogenous homeostasis of porphyrins, particularly with the regulation of the transport of protoporphyrin IX and other intermediates in the biosynthesis of the heme group [13–16].
In rodents, the Harderian gland (HG) has been reported to be an intraorbital structure that plays an important role in the biosynthesis and storage of porphyrins. Specifically, in the HG of the female Syrian hamster, the porphyrinogenic enzymatic activity is higher than that in the HG of the male hamster [17–19]. This porphyrin biosynthetic pathway, in addition to fatty acids, indoleamines, and somatostatin biosynthesis, exhibits sex differences that are associated with the levels of sex steroids and their nuclear receptors [20–24]. Gonadectomy in male hamsters has been observed to suppress this sexual dimorphism and feminize the male gland, whereas the administration of androgens (such as testosterone and dihydrotestosterone) may prevent this glandular feminization. However, it has been reported that ovariectomy does not cause the masculinization of HG and its effects on the activity and structure of HG are less dramatic; however, when testosterone is administered to females, HG acquires the characteristics of the male gland; these data have been suggested to be an androgenic control in porphyrin metabolism [17,24].
Previously, we have shown that the ABCB6 transporter has little association with the Harderian porphyrins and lacks a sexual dimorphism [25]. Because the hamster HG displays a marked sexual dimorphism in the production and storage of large amounts of protoporphyrins, in the present study, the cloning and endocrine regulation of the hamster ABCG2 transporter was analyzed and reported to establish whether the ABCG2 transporter is associated with sexual dimorphism and endogenous porphyrin homeostasis.
2 Material and methods
2.1 Animals
This study was performed using female (n = 45) and male (n = 20) adult Syrian hamsters (Mesocricetus auratus), which were housed under 12 h/12 h light/dark cycle conditions with water and fed ad libitum. Twenty-four hours after gonadectomy, the subgroups were treated in vivo with a single daily dose of each steroid. The hamsters were subdivided into different experimental groups according to the physiological conditions that were previously mentioned [25].
The experimental design was as follows: an intact male group, a bilaterally orchidectomized group for 7 days, a bilaterally orchidectomized group treated daily with testosterone (1 mg) supplementation for 6 days, and a bilaterally orchidectomized group treated daily with vehicle (50 μl of corn oil) supplementation for 6 days.
In females, the stages of the estrous cycle [proestrus (P), estrus (E), metestrus (M), or diestrus (D)] were determined by vaginal smears. One group was bilaterally ovariectomized for 7 days, three groups were bilaterally ovariectomized for 7 days and received the following treated daily for 6 days: progesterone (1 mg), 17β-estradiol (10 μg), progesterone plus 17β-estradiol (1 mg plus 10 μg), or vehicle (50 μl of corn oil).
The hamsters were anaesthetized with ketamine:xylazine (80 mg/kg:8 mg/kg, intramuscularly) before gonadectomy and decapitated 24 h after the last injection. In males, a 1.5-cm incision was made at the scrotum. The testes were exposed, ligated, extracted, and then the wound sutured closed. The females were ovariectomized by making a 1.0-cm incision medial to the flank glands. The uterine horns were ligated and the ovary extracted. The muscles and skin were sutured shut separately. Different tissues were immediately removed, frozen on dry ice, and stored at −70 °C until the assays were performed.
Here, we only utilized the HGs of males and females under different endocrine conditions to determine the sex steroids’ impacts on ABCG2 expression and to illustrate the sexually dimorphic pattern in the HGs. Only for the expression comparative aims with the hamster HG; the cerebellum, ovary, uterus, adrenals, testis, liver, epididymis, spleen, brain, gut, heart, kidney, hypothalamus, seminal vesicles, and lungs were obtained. The Ethics Committee for Research in Animals at our institution (INCMNSZ, BRE-1930-18-19-1) approved the research for the care and use of animals.
2.2 RNA extraction and cDNA synthesis
Total RNA from the HGs and peripheral tissues was isolated using the TRizol technique (Invitrogen, Carlsbad, CA, United States), according to the manufacturer's instructions. The purity and concentration values (at 260/280 nm) of the total RNA samples were measured using a spectrophotometer. The total RNA (20 μg) integrity was checked using 1% agarose gel electrophoresis in each experimental assays and verified by the presence of ribosomal RNAs. Total RNA samples were treated with DNase (Promega Corporation, Madison, WI, United States). Total RNA (1 μg) was reverse-transcribed (RT) to complementary DNA (cDNA), according to the manufacturer's instructions (Transcriptor First Strand cDNA Synthesis Kit, Roche Diagnostics, IN, United States). All the cDNAs were stored at −20 °C until they were used.
2.3 Isolation and molecular cloning of ABCG2 cDNA
The 5′ and 3′ ends of the ABCG2 cDNAs were amplified using SMART Rapid Amplification of cDNA Ends (RACE) kit (Clontech, Mountain View, CA, United States) according to the manufacturer's instructions. Two degenerate primers (sense 5ʹ-GAAAGAT/CCCAA/CA/GGGGATTATCTG-3ʹ and antisense 5ʹ-CCGTCT/CTCTTCAGTCCTAAC-3ʹ) and two gene-specific primers (5ʹ-GAACCACATAGCCTGAAGTA-3ʹ and 5ʹ-CTTACTTCTTTGGAAAGCTGAT-3ʹ) were designed for the RACE assays. Degenerate primers were designed from the nucleotide sequences of mouse (NM_001355477) and rat (NM_181381) ABCG2. The specific primers for amplifying the complete ABCG2 cDNA were: sense 5ʹ-GGAAGGCAGAAATCTGAAA-3ʹ and antisense 5ʹ-AGGATTCAAGTAATAGTAGGGTAAAGG-3ʹ. The full-length cDNA was cloned using the pcDNA3.1/CT-GFP-TOPO TA vector (Invitrogen Co., Carlsbad, CA, United States). Plasmid DNA, which contained full-length ABCG2 cDNA, was purified using the GenElute Five-Minute Plasmid Miniprep kit (Sigma-Aldrich, St. Louis, MO, United States) and PureYield Plasmid Maxiprep kit (Promega, Woods Hole, WI, United States), according to the manufacturer's protocols.
2.4 Sequencing of ABCG2 and cDNA/protein analysis
From hamster HG cDNA, full-length cDNA sequencing of ABCG2 was performed in the sense and antisense directions from at least four different clones of each cDNA using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Austin, TX, United States) on an ABI-PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA, United States). All sequences were confirmed in both directions. The Open Reading Frame (ORF) in the ABCG2 sequence was determined using the ORF Finder (http://www.ncbi.nlm.nih.gov/orffinder/). The deduced amino acid sequence of the ABCG2 polypeptide was performed through the Expert Protein Analysis System (http://www.expasy.org/), and the physicochemical properties were determined using ProtParam (https://www.web.expasy.org/protparam/). The prediction of subcellular localization was determined by DeepLoc (http://www.cbs.dtu.dk/services/DeepLoc). The transmembrane regions were identified using TMHMM v.2.0 Server (http://www.cbs.dtu.dk/services/TMHMM/). Multiple alignments of hamster ABCG2 (AMA11217.1) and other vertebrate G2 transporters (human: NP_004818.2; mouse: NP_036050.1; rat: NP_852046.1) were constructed to analyze the conserved motifs using the CLUSTALW software (http://www.genome.jp/tools/clustalw/). Sequencing of the cloned cDNA was performed using the BLAST program and the identity percentage was determined (http://www.ncbi.nlm.nih.gov/BLAST/). Nucleotide (accession No. KU237247) and amino acid (accession No. AMA11217.1) sequences were deposited and submitted to GenBank (accession No. KU237247).
2.5 Construction of the ABCG2 phylogenetic tree
We downloaded ABCG2 proteins that were already reported in different species from the NCBI (https://www.ncbi.nlm.nih.gov/protein) database. The downloaded amino acid sequences were aligned with the multiple sequence alignment tool CLUSTALW (https://www.genome.jp/tools-bin/clustalw). The multiple alignment formats were obtained in FASTA file format using the protein database for ABCG2 (https://www.ncbi.nlm.nih.gov/protein/). Phylogenetic trees were visualized using Molecular Evolutionary Genetics Analysis X (MEGA X) including 43 proteins from mammalian species.
2.6 Modelling 3D structure
The 3D structure of the ABCG2 protein was generated by using template-based modelling with the intensive mode of the Phyre2 software [26]. The transmembrane helices of the hamster G2 transporter were predicted in the amino acid sequence.
2.7 Gene expression quantification
The total RNA was isolated using the TRizol method followed by DNase treatment. RT-qPCR reactions were performed using the LightCycler® 480 Probes Master kit (Roche Diagnostics, IN, United States) in a final volume of 20 μL for each biological tissue sample. The assays were optimized using 10 μL of TaqMan Master LightCycler 2×, 0.5 μM final concentration of each sense 5ʹ-ACTGCGAGCCCTACAATAAC-3ʹ and antisense 5ʹ-CCTGCTCTCCTTCCTCTCTAT-3ʹ (amplicon size of 96 base pairs [bp]) primers, 0.4 μL of universal probe (ABCG2 and β-actin probe #9 [04–685–075–001], Germany Roche Diagnostics, Mannheim, Germany), and 5 μL of cDNA (5 ng cDNA/well). The thermal cycling conditions were as follows: one cycle of pre-incubation at 95 °C for 10 min, 40 cycles of amplification at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 1 s, with a final cycle of cooling at 40 °C. The quantitative expression of ABCG2 was measured using the LightCycler 480 II instrument (Roche Life Science, IN, United States), and the results were analyzed using the relative quantification method provided by the LightCycler software (Version 4.5). The relative levels of ABCG2 mRNA were normalized to hamster β-actin mRNA levels (GenBank accession No. AJ312092; sense: 5ʹ-AGCTATGAGCTGCCTGATGG-3ʹ and antisense: 5ʹ-CAGGAAGGAAGGCTGGAAA-3ʹ; amplicon size of 87 bp). qPCR assays for each sample were performed five times independently. The relative expression results are presented as the mean ± standard deviation (SD) of five independent qPCR runs for each group. The methods of statistical analysis from each hormonal condition and multiple comparisons were assessed using Dunnett's test [27]. Differences were considered significant at P < 0.05.
3 Results
3.1 cDNA cloning and sequencing of hamster ABCG2
We isolated and determined the full-length cDNA sequence of hamster ABCG2 using the RACE-PCR procedure. The cloned cDNA (2098-bp) contains a 1971-bp ORF (GenBank accession number: KU237247) that encodes a protein with 656 amino acids (GenBank accession number: AMA11217.1). ABCG2 cDNA begins with the typical ATG codon and ends with a TAA codon. The 5ʹ-untranslated region (5ʹ-UTR) of ABCG2 cDNA consists of 58 bp, while the 3ʹ-UTR contains 57 bp that includes an ATTAAA alternative polyadenylation signal and a portion of the poly(A) tail localized 19 bp downstream of the hexamer sequence consensus (Fig. 1). The structural analysis of the ABCG2 transporter revealed a high percentage of similarity between vertebrate species (Table 1), which suggests that the gene is conserved along the phylogeny.
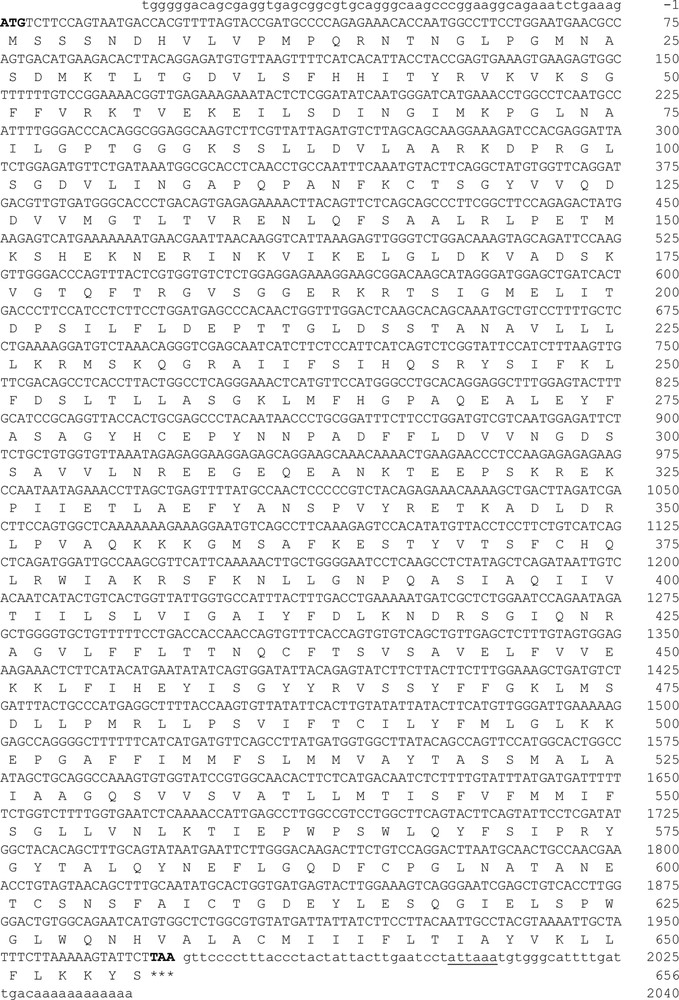
Full-length cDNA of ABCG2 (GenBank accession No. KU237247). Nucleotide and deduced amino acid sequence of the hamster ABCG2 transporter. In the sequence, the start codon (ATG) is indicated by the number 1 and stop codon (TAA) is indicated by three asterisks. The polyadenylation signal sequence is underlined (ATTAAA).
Comparative analysis of the functional domains of hamster ABCG2 with other vertebrate transporters.
| Species | #a.a. | % nt | % aa | # access | Walker A Domain | Walker B Domain | Q, D, H-loop Domains | C-Signature |
| Hamster | 656 | 100 | 100 | AMA11217 | 100 | 100 | 100 | 100 |
| Rat | 657 | 87 | 88 | NP_852046.1 | 100 | 100 | 100 | 90 |
| Mouse | 657 | 88 | 89 | NP_036050.1 | 100 | 100 | 100 | 90 |
| Human | 655 | 81 | 84 | NP_004818.2 | 100 | 100 | 100 | 100 |
3.2 Analysis of ABCG2 polypeptide
The calculated molecular mass of ABCG2 was 72.844 kDa with a theoretical isoelectric point (pI) of 8.61. The aliphatic index and the grand average of hydropathicity (GRAVY) were 94.98 and 0.094, respectively. The prediction of protein subcellular localization was identified in the lysosome/vacuole (likelihood 0.47590) and membrane (likelihood 1). Multiple alignment analysis of deduced hamster ABCG2 amino acid sequences with other vertebrates displayed several conserved domains. Six putative TMDs (393–415, 428–450, 477–496, 506–528, 535–557, and 630–652 amino acids) and four Extracellular Loops (ECL1-4) inferred from TMHMM Server and Phyre2 were identified from hamster ABCG2 protein. In the NH2-terminal region, Walker A (78–85 amino acids), Walker B (204–209 amino acids), and C signature (184–193 amino acids) motifs were localized by sequence comparative alignment (Fig. 2). The consensus sequences Q-loop (121–124 amino acids), D-loop (212–215 amino acids), and H-loop (237–243 amino acids) were identified and presented 100% homology. In ABCG2 polypeptide, we identified three conserved cysteine residues located at positions C591, C602, and C609 of the C-terminal region that were involved in dimerization. Sequence similarity analysis by BLAST and CLUSTALW programs showed that the hamster ABCG2 protein is conserved phylogenetically within the vertebrate hemi-transporters and that it shares a high percentage of identity with the mouse (89%, GenBank accession number: NP_036050), rat (88%, GenBank accession number: NP_852046.1), and human (84%, GenBank accession number: NP_004818.2) hemi-transporters (Table 1). The predicted topological structure of hamster ABCG2 is described in Fig. 3.
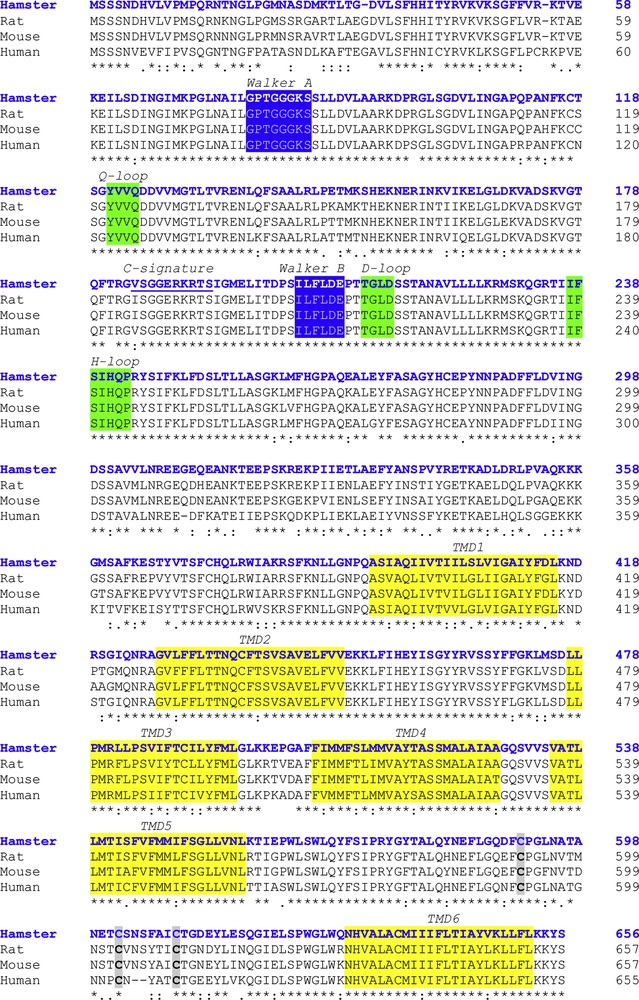
Multiple sequence alignment of the ABCG2 of hamster (blue colored font) with other vertebrate transporters. The Walker domains are indicated in the blue boxes. The consensus sequences Q-loop, D-loop, and H-loop are shown in the green boxes. Identical amino acid residues are marked with asterisks. The TMDs are shown in the yellow boxes. The three cysteine residues located at positions C591, C602, and C609 are indicated in grey boxes. The C-signature is underlined.
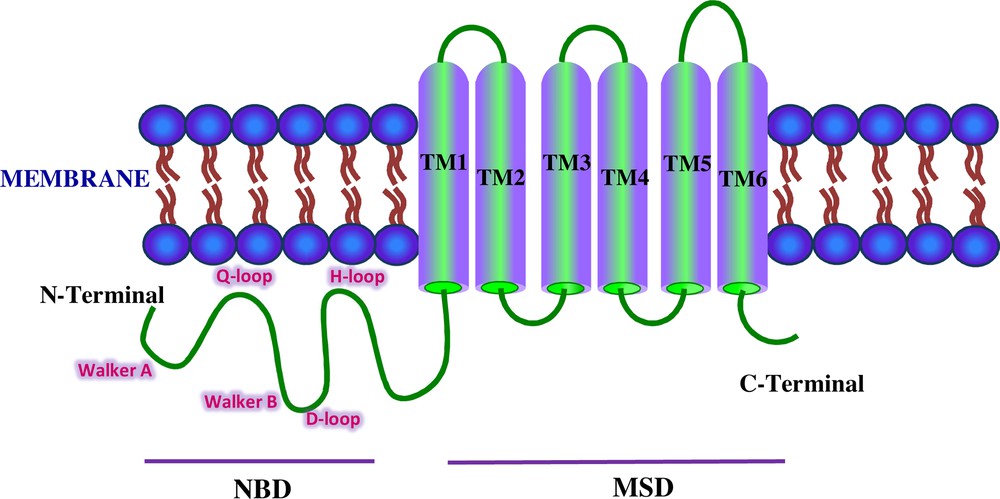
Schematic representation of the ABCG2 transporter. The domains shown are NBD and MSD. The Walker A, Walker B, and C signature motifs are localized in the NH2-terminal region. The Q-loop, D-loop, and H-loop are shown in pink colored font.
3.3 Protein phylogenetic tree analysis
Phylogenetic tree analysis and amino acid sequence comparison among these 43 proteins belong to seven clades (I–VII) indicating that the structural similarities that are shared by mammalian species are probably a result of ABCG2 gene duplication. Five of them are grouped in primates (I), carnivora (II), cetacea (IV), rodentia (V), and artiodactyla (VII) (Fig. 4).
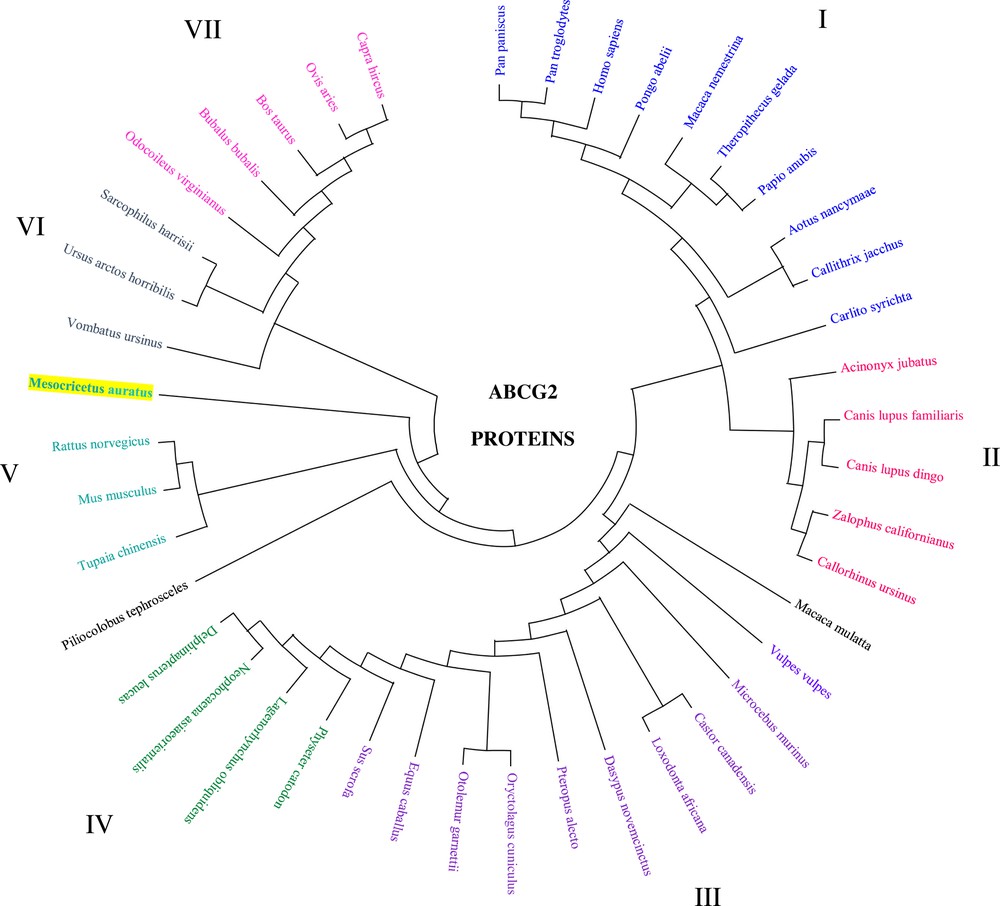
Phylogenetic tree analysis of the ABCG2 amino acid sequence in the hamster and other mammalian species. Mammalian clusters that were manually selected based on the amino acid tree are illustrated in different colors. The evolutionary history was inferred using the Minimum Evolution (ME) method. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1. The Neighbor-Joining algorithm was used to generate the initial tree. This analysis involved 43 amino acid sequences. Evolutionary analyses were conducted in MEGA X. (GenBank accession number: NP_004818.2, NP_036050.1, NP_852046.1, NP_001032555.2, NP_001041486.1, NP_001072125.1, NP_001028091.1, NP_001272636.1, AMA11217.1, NP_001316001.1, NP_999175.1, JAV44144.1, XP_027709188.1, XP_027625661.1, XP_027455628.1, XP_026972704.1, XP_014929119.1, NP_001277803.1, XP_026365535.1, XP_023053858.2, XP_025853897.1, XP_025729196.1, XP_025281012.1, XP_025241791.1, XP_024901778.1, XP_008961902.1, XP_011736036.1, XP_024591860.1, XP_024211938.1, XP_024102041.1, XP_007102590.1, XP_023493515.1, XP_004463978.1, XP_023409034.1, XP_023369130.1, XP_012408409.1, XP_022438006.1, XP_009205420.1, XP_008055687.1, XP_021528929.1, XP_020724633.1, XP_020140157.1, and JAB42602.1).
3.4 Structure of ABCG2
The 3D model of hamster ABCG2 (Fig. 5) was generated with 100% confidence and coverage of 85% of residues (560 amino acids). In the C-terminal region, six transmembrane α-helices were localized. Topologically, the model predicted four ECL and three cytoplasmic loops.

Predicted 3D structure of ABCG2 transporter in the hamster. The model aligned 560 residues and predicted by with 100% confidence, 85% identity, and 3.78 Å resolution.
3.5 Tissue distribution and sex steroid-dependent expression of ABCG2 mRNA
To investigate the distribution and expression profiles of ABCG2 in the Syrian hamster, qPCR reactions were performed to evaluate the mRNA levels. The transcript was detected in all hamster tissues that were tested. The highest expression levels were found in the testis, ovary, gut, brain, kidney, cerebellum, and HG. The lowest expression was detected in the adrenals, liver, heart, and spleen (Fig. 6). In HG, ABCG2 gene expression exhibits a sexually dimorphic pattern where females show higher mRNA levels than males. The results revealed that androgen deficiency in the castrated group could induce ABCG2 mRNA expression, whereas replacement with testosterone in the castrated group could restore mRNA expression. The data revealed significant differences in ABCG2 mRNA expression levels during the estrous cycle and after gonadectomy in females, where the expression of ABCG2 was upregulated. The high ABCG2 expression levels in the estrous cycle were identified in metestrus. The administration of progesterone and/or 17β-estradiol to the group of ovariectomized females restored the levels of ABCG2 gene expression. The data showed that ABCG2 expression is regulated by sex steroid levels (Fig. 6).
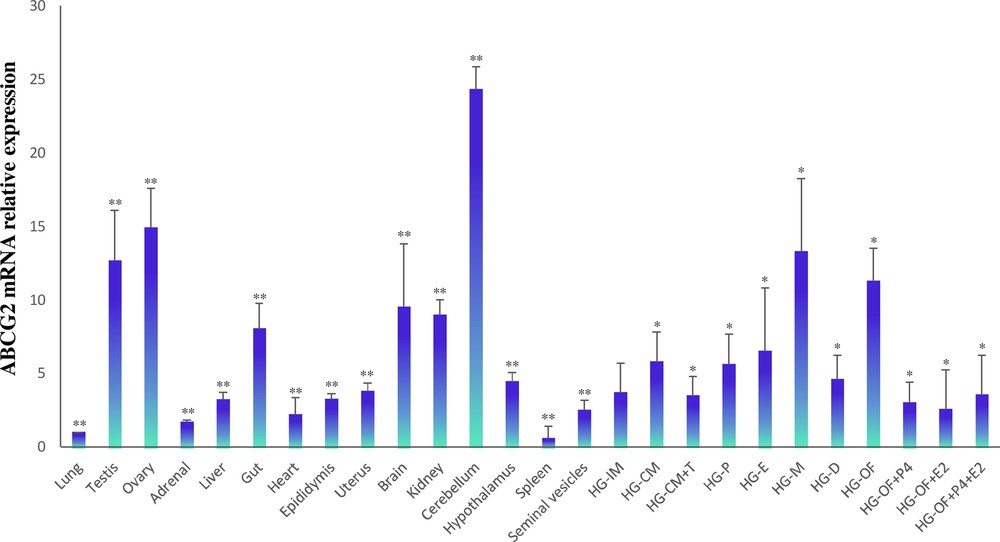
Relative ABCG2 expression levels analyzed by RT-qPCR in different hamster tissues. The impact of sex steroids on ABCG2 mRNA levels in the hamster HG was measured. HG-IM, Harderian gland of the intact male; HG-CM, HG of a castrated male; HG-CM + T, HG of a castrated male plus testosterone; HG-P, HG in proestrus; HG-E, HG in estrus; HG-M, HG in metestrus; HG-D, HG in diestrus; HG–OF, HG of ovariectomized female; HG–OF + P4, HG of ovariectomized female plus progesterone; HG–OF + E2, HG of ovariectomized female plus 17β-estradiol; HG–OF + P4 + E2, HG of ovariectomized female plus progesterone and 17β-estradiol. Results were normalized to β-actin. Data are presented as the mean ± SD, with n = 5 biological independent replicates. The star (*) indicates the statistical significance between HG–MI and HGs of males and females under different endocrine conditions (P < .05); **represents the statistical significance between HG–MI and different tissues (P < 0.05).
4 Discussion
The ABC superfamily functions as intracellular transporters of inorganic and organic molecules that bind and hydrolyze ATP. The ABCG2 transmembrane transporter has an important function in the xenobiotic protection and drug transport inside the blood–brain barrier, the blood–testis barrier, and the maternal–fetal barrier [9,28]. Similarly, it has been observed that the ABCG2 transporter displays an important contribution in regulating intracellular porphyrin efflux and heme homeostasis [16]. In the HG of some rodents, specifically the Syrian hamster, the amount of porphyrins is elevated and it showed a sexual dimorphism that was dependent on sex steroids [29]; however, the association between porphyrin metabolism and ABC transporters requires further information [25]. Therefore, in this study, the structure and physiological functions of the porphyrin ABCG2 transporter in this intraorbital gland were elucidated. We consider that isolation and molecular cloning of the ABCG2 cDNA subfamily provides a foundation for analyzing its expression and function.
In the current study, the full-length cDNA of ABCG2 from the Syrian hamster was isolated and characterized. For the NH2-terminal region, we identified the Walker A (78–85 residues; G-X-X-G-X-G-K-S), Walker B (204–209 residues; I-L-F-L-D-E), and C signature (184–193 residues; V-S-G-G-E-R-K-R-T-S) motifs within its predicted amino acids sequence, as well as characteristic elements of ABC transporters and highly conserved regions of NBD or ABC (Table 1). Based on the molecular structure and comparative analysis of the cloned ABCG2, we showed isolated cDNA codes for a protein belonging to the ABC transporters superfamily in hamsters. Other key structural regions, including the Q-loop, D-loop, and H-loop, were identified in the primary structure of the hamster G2 transporter. These loops have been associated with ATP hydrolysis and with the coupling between NBD and TMD. Analysis of the amino acid sequence suggests that the hamster ABCG2 polypeptide has all the fundamental structures for transport activity.
To date, it has been reported that the half-transporter ABCG2 or BCRP1 contains six transmembrane α-helices that are localized in the C-terminal region. These helices play an essential role in determining overall transport activity and substrate specificity [6,30,31]. In the hamster ABCG2 transporter, only one TMD with six transmembrane helices and four ECLs were determined. We identified three cysteine residues that were highly conserved in the C-terminal region and have been associated with the dimer/oligomer formation through intermolecular disulfide bond formation. Similarly, mutagenesis assays in the TMD3 have identified two amino acid residues (Arg482 and Pro485) that play an essential role in substrate specificity and/or overall transport activity; these amino acids have a high homology percentage (100%) between the hamster and other vertebrate species [28,32]. Taken together, the results indicated that the cloned sequence of hamster ABCG2 could be functional structurally in the HG, and according to their high percentage of homology, the hamster ABCG2 gene appears to be well conserved throughout evolution. Thus, we concluded that ABCG2 isolated in this study is a functional transporter ABC superfamily.
The phylogenetic analyses showed the presence of seven clades (I–VII), five of which are clearly delimited and grouped by similar characteristics (I, II, IV, V, and VII); however, in two clades (III and VI), a phylogenetic heterogeneity is revealed and a different grouped species can be observed. This type of interpretation could result from our lack of knowledge about the gene and amino acid sequences of the ABCG2 transporters, so we suggest that these clades are incomplete and the exact and precise cloning and sequencing of more ABCG2 genes is required to establish the phylogenetic relationships in the mammalia class and in other chordates.
In human tissues, the distribution of the ABCG2 protein has been shown in the small intestine, liver, kidney, and blood–brain and blood–placental barriers [33], suggesting that ABCG2, which is also known as BCRP1, contributes to the absorption, elimination, and distribution of biomolecules or xenobiotics, principally for drug transport. In addition, several studies have reported that ABCG2 is associated with porphyrin homeostasis and heme transportation [14,16]. Our data performed in the Syrian hamster revealed that ABCG2 was expressed in different tissues, including the testis, epididymis, and porphyrinogenic tissues such as the liver and adrenal glands; in the HG, the highest expression levels were detected in the Harderian tissue in females and it showed variations during the estrous cycle, specifically in metestrus. The data suggest that the G2 transporter is likely involved in the endogenous regulation of female porphyrins or heme in this porphyrinogenic tissue. Therefore, the physiological role of ABCG2 is likely to be closely related to the specific distribution profile of ABCG2 detected in our study.
The present study demonstrates that the ABCG2 mRNA tissue distribution exhibited a marked sexual dimorphism in the HGs. mRNA expression levels were lower in intact male HG, whereas the expression levels were higher during metestrus. These differences, which were observed in the mRNA expression profile between males and females, suggest that ABCG2 is under the control of gonadal steroids. To obtain more evidence on this, hamster HGs were treated and analyzed under different hormonal conditions. We demonstrated that the castrated males showed less drastic effects compared with the ovariectomized females; our results show that ovariectomy induces an increase in ABCG2 transcript expression, suggesting steroid-regulated ovarian control of the G2 transporter and that the high concentrations of porphyrins in female HG are likely controlled by ABCG2 expression in this tissue. Our results are in agreement with the characteristics of sex steroids in the potential control of Harderoporphyrin movement and biosynthesis. Previous investigations have reported and discussed the effects of sex steroids on the anatomical and biochemical changes in the HG [29,34–37]. Thus, several lines of evidence have reported that the administration of androgens to females decreases the porphyrinogenic activity, while in the ovariectomized females, the total porphyrin content remains static and does not result in the masculinization of the female HG. However, the enzyme activity for the porphyrin biosynthesis is markedly reduced in ovariectomized females, which suggests gonadal hormone control [38,39].
5 Conclusion
In summary, the data presented in this study suggest that the ABCG2 that was isolated has a structure and properties that are consistent with a member of the ABC transporter family. Similarly, we determined distinct patterns of ABCG2 transcriptional profiles, and specifically, a sexually dimorphic expression profile was observed in HG. In this gland, the control of ABCG2 expression levels appears to be regulated by gonadal sex steroids. Our findings suggest that ABCG2 might be an essential transporter in porphyrin metabolism in this intraorbital tissue.
Author contributions
L.M., F.V., and L.R. coordinated the research project. B.C. and L.M. performed the experiments. F.V. and L.R. analyzed the data and wrote the paper. All authors have approved the manuscript for publication.
Disclosure of interest
The authors declare that they have no competing interest.


