1. Introduction
The relationship between genetic diversity and the invasiveness of alien species has always been a hot issue in invasive ecology [1]. Most studies showed that the level of genetic diversity of an alien species was positively correlated with its invasiveness [2], which is important for the successful invasion and rapid population expansion of an alien species [3]. However, some researchers have found that some noxious invasive species do not have high genetic diversity in the invaded areas [4, 5]. The authors hold the view that low genetic diversity caused by cloning reproduction and selfing events maintains the fitness of an alien species in the introduced habitats [6, 7, 8]. Therefore, more evidence is needed to reveal the mechanism of the successful invasion of an alien species from the perspective of population genetics [9].
Xanthium orientale subsp. italicum (Moretti) Greuter, a native plant of North America belonging to the Asteraceae family, is now widely distributed in many areas of Europe, Asia, and Oceania [10], posing a serious threat to agricultural production and biodiversity conservation in the introduced areas [11]. After the discovery of the first X. italicum tree in Changping District, Beijing, China in September 1991 [12], the distribution area and expansion rate of X. italicum in China sharply increased in the past 28 years. X. italicum has spread to six provinces/municipalities in China, including Beijing, Hebei, Shandong, Liaoning, Shaanxi, and Xinjiang [13], from the eastern (Shenyang and Liaoning Province) to the western (Yining and Xinjiang Province) provinces. The farthest spatial distance was more than 5000 km. As a common species in the habitat of origin, it can expand arbitrarily in the invaded land, efficiently crowding out the local species and encroaching on the whole ecosystem. However, the invasion mechanism of X. italicum is still unclear. To elucidate this mechanism, many researches have been carried out and many hypotheses have been put forward. These hypotheses include the “inherent superiority” hypothesis, in which the success of some invasive alien plants is due to its unique biological features or intrinsic advantages (such as morphology, ecology, physiology, and genetic features). Compared with indigenous species, alien species with intrinsic advantages may have more genetic variation in evolution, forming ecotypes that are more suitable for a wide range of environmental conditions and utilising more resources, or have stronger ability or character to resist external environmental stress, so as to eventually gain an advantage in a competition or occupy a niche that some indigenous species cannot use, thus successfully invading the area [14]. Despite studies on the ecological adaptability of the seeds [15], interspecific competitiveness [16], reproductive biological characteristics [17], and allelopathic potential [13] of X. italicum, our understanding of its successful invasion mechanism is still not sufficiently comprehensive.
Geographical distribution of the 10 sampled populations of X. italicum in China. Sampled populations are indicated with filled dots.
A molecular marker is an important and useful tool in molecular biology and can be used in studies of invasive species as a mean to identify source populations, determine invasion routes, and elucidate invasion mechanisms and success [18]. Obviously, genetic variability evaluation of X. italicum in invasive ranges deserves more attention. Therefore, we collected the leaves of X. italicum from 10 natural populations in China and analysed its genetic diversity and genetic differentiation pattern by using the inter-simple sequence repeat (ISSR) molecular marker technique. The purpose of this study was to reveal the level of genetic diversity, degree of genetic differentiation, and gene flow of X. italicum populations, and to analyse its genetic structure, so as to provide a theory basis for the invasion mechanism of this invader.
2. Materials and methods
2.1. Plant materials
A total of 185 individuals, which corresponded to 10 populations, were sampled across its entire geographic range in China, including Beijing, Liaoning, Hebei, Shandong, Shaanxi, and Xinjiang Province (Figure 1). The sampling was conducted according to the methods described by Roose et al. [19]. Three fresh healthy leaves were randomly collected from one tree, and three other leaves from a different tree were selected at a standard interval of 10 m. Fresh leaves were dried in a Ziplock bag with silica gel and then transported back to the laboratory. Corresponding to each population, parameters such as longitude, latitude, and altitude were recorded for further analyses (Table 1).
Geographic localities and sample sizes of X. italicum populations
| Population | ID | Location | Sample | Longitude (E) | Latitude (N) | Altitude (m) |
|---|---|---|---|---|---|---|
| YL | 1 | Xinjiang Province | 25 | 82.87 | 43.57 | 887 |
| SHZ | 2 | Xinjiang Province | 20 | 86.16 | 44.54 | 453 |
| XA | 3 | Shanxi Province | 12 | 109.07 | 34.31 | 387 |
| GT | 4 | Hebei Province | 26 | 115.59 | 40.26 | 483 |
| DWC | 5 | Beijing City | 16 | 116.27 | 40.17 | 42 |
| GHC | 6 | Beijing City | 20 | 116.27 | 40.17 | 121 |
| JZ | 7 | Liaoning Province | 20 | 121.04 | 41.13 | 40 |
| LTS | 8 | Liaoning Province | 20 | 121.20 | 38.75 | 34 |
| WLH | 9 | Liaoning Province | 11 | 123.44 | 41.75 | 46 |
| SD | 10 | Shandong Province | 15 | 122.06 | 37.53 | 40 |
2.2. DNA extraction and polymerase chain reaction (PCR) amplification
Total genomic DNA was extracted by using a DNA quick plant system Kit (Beijing, China), and then dissolved in 0.1× TE buffer. The ISSR primers were synthesised by Shanghai Sangon Biological Engineering Technology & Service Co., Ltd. (China) corresponding to the primer set (#9) published by the University of British Columbia (Vancouver, British Columbia, Canada) [20].
The ISSR primers used in this study
| Primer | Sequence R = (A, G), Y = (C, T) | Annealing temperature (°C) |
|---|---|---|
| UBC810 | GAGAGAGAGAGAGAGAT | 44.9 |
| UBC812 | GAGAGAGAGAGAGAGAA | 45.1 |
| UBC834 | AGAGAGAGAGAGAGAGYT | 48.7 |
| UBC835 | AGAGAGAGAGAGAGAGYC | 50.2 |
| UBC836 | AGAGAGAGAGAGAGAGYA | 48.4 |
| UBC840 | GAGAGAGAGAGAGAGAYT | 47.0 |
| UBC868 | GAAGAAGAAGAAGAAGAA | 41.8 |
| UBC880 | GGAGAGGAGAGGAGA | 47.9 |
PCR was performed in a 25-μL reaction volume containing 2× Taq PCR MasterMix, 10 μM primer, and 60 ng DNA template. The amplifications were performed in a thermal cycler (Eppendorf, Germany) with the following program: initial denaturation at 94 °C for 5 min; 40 cycles of 94 °C for 45 s, an appropriate annealing appropriate temperature (Table 2) for 45 s, 72 °C for 1.5 min; and the last extension at 72 °C for 10 min. A negative control with no DNA was included in each PCR run.
The amplification products were separated on 1.5% agarose gels (1× TAE buffer) at 120 V for 0.5 h, stained with ethidium bromide (0.5 mg/ml), and visualised in ultraviolet light by using an automatic Gel Documentation System (Syngene, USA). DL2000 ladder (Shanghai Sangon Biological Engineering Technology & Service Co., Ltd.) was applied as a marker of DNA molecular weight [21].
2.3. Statistical analyses
As ISSR markers are dominantly inherited, each band was assumed to represent the phenotype at a single biallelic locus [22]. Only bright and discernible bands ranging from 200 to 2000 bp were considered in the final statistical analyses. A binary data matrix was built by scoring ISSR bands as presence (1) or absence (0) characters.
Estimations of genetic diversity within and among populations were obtained with the POPGENE software version 1.31 [23]. We calculated the percentage of polymorphic loci (PPL), Nei’s gene diversity (H), and Shannon’s information index (I). For evaluation of population genetic differentiation, total genetic diversity (Ht), mean intra-population genetic diversity (Hs), coefficient of genetic differentiation (GST), and gene flow (Nm) of 10 populations were computed. Analysis of molecular variance (AMOVA) [24] was employed to statistically assess the partitioning of genetic variability within and among populations using the DCFA1.1 program [25].
An unweighted pair group method with the arithmetic average (UPGMA) dendrogram was obtained using the NTSYS-pc 2.11 software [26]. A Mantel test [27] of genetic distances and geographic distances was conducted by using the software package GenAlEx v6.3 [28] and TFPGA v1.3 [29].
3. Results
3.1. Polymorphism analysis and genetic diversity of X. italicum
Eight ISSR primers yielded a total of 76 clear bands in the 185 individuals from 10 populations, and all bands were polymorphic (100%). The number of bands varied from 8 to 12, with an average of 9.5 bands per primer. Pop4 has the most PPL (93.42%), whereas Pop10 produced the least PPL (14.47%) among the 10 populations. The highest H was that of Pop4 (0.3234), whereas Pop10 showed the lowest H (0.0564). Moreover, other indicators of the variation trend were consistent with the PPL. The values of Ne and I varied from 1.0970 to 1.5650 and from 0.0832 to 0.4812, respectively. The X. italicum populations had relatively lower diversity at the population level (PPL = 60.26%; Ne = 1.3621; H = 0.2098; I = 0.3129), but had significantly higher genetic diversity at the species level (PPL = 100.00%; Ne = 1.6425; H = 0.3673; I = 0.5425) (Table 3).
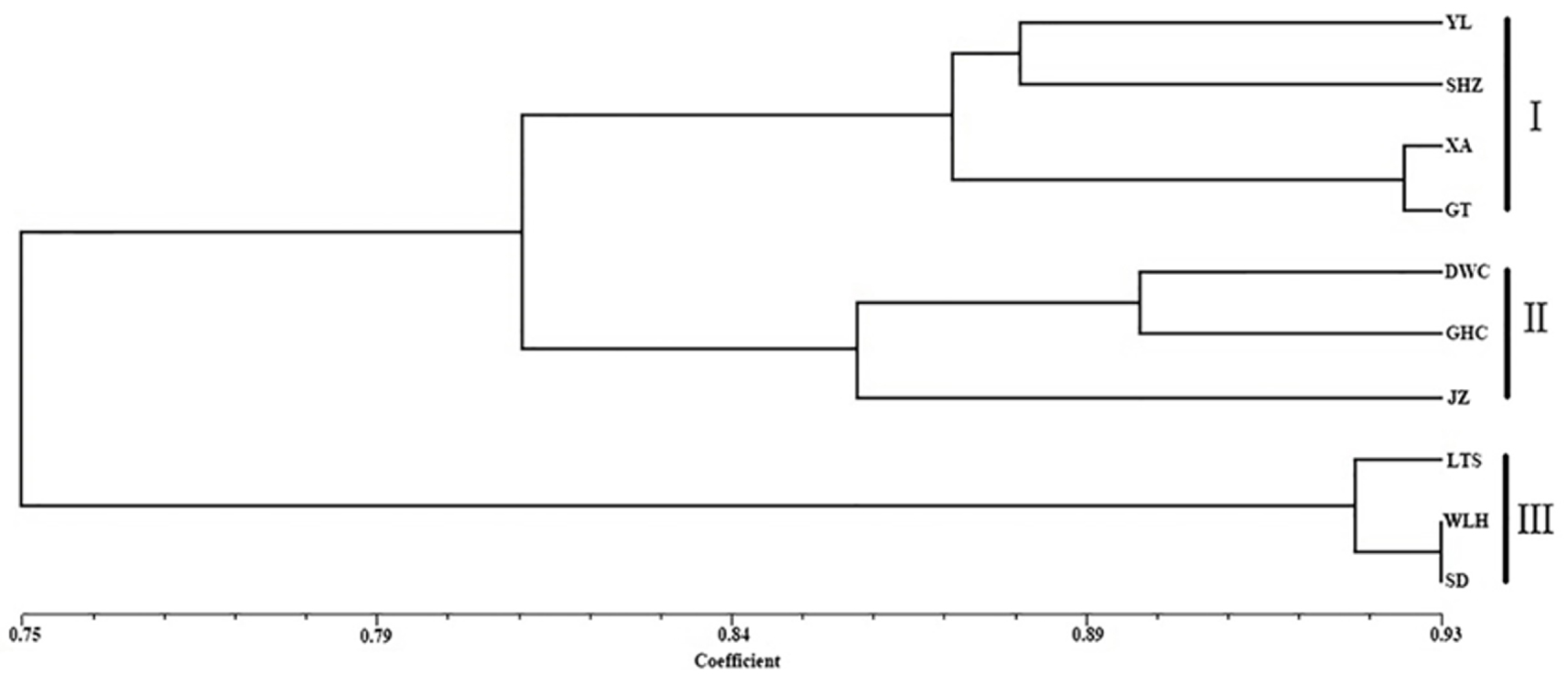
UPGMA dendrogram based on Nei’s (1978) genetic distances among X. italicum populations.
Genetic diversity of 10 X. italicum populations
| Population | Na | Ne | H | I | PPL (%) |
|---|---|---|---|---|---|
| Pop1 | 1.8947 | 1.5611 | 0.3157 | 0.4667 | 89.47 |
| Pop2 | 1.8553 | 1.4560 | 0.2767 | 0.4204 | 85.53 |
| Pop3 | 1.7500 | 1.4607 | 0.2687 | 0.4001 | 75.00 |
| Pop4 | 1.9342 | 1.5650 | 0.3234 | 0.4812 | 93.42 |
| Pop5 | 1.5658 | 1.3461 | 0.1993 | 0.2964 | 56.58 |
| Pop6 | 1.4868 | 1.3048 | 0.1770 | 0.2625 | 48.68 |
| Pop7 | 1.7105 | 1.4691 | 0.2623 | 0.3862 | 71.05 |
| Pop8 | 1.3816 | 1.2006 | 0.1201 | 0.1830 | 38.16 |
| Pop9 | 1.3026 | 1.1610 | 0.0980 | 0.1490 | 30.26 |
| Pop10 | 1.1447 | 1.0970 | 0.0564 | 0.0832 | 14.47 |
| Population level | 1.6026 | 1.3621 | 0.2098 | 0.3129 | 60.26 |
| Species level | 2.0000 | 1.6425 | 0.3673 | 0.5425 | 100.00 |
Na: observed number of alleles; Ne: effective number of alleles; H: Nei’s (1973) gene diversity; I: Shannon’s information index; PPL: percentage of polymorphic loci.
3.2. Genetic differentiation and gene flow in X. italicum populations
Genetic differentiation of X. italicum populations, as calculated by AMOVA (Table 4), showed that 22.72% of the total variation was attributed to among populations and 77.28% of the total variation originated from within populations. Both showed a highly significant (P < 0.001) level. The coefficient of gene differentiation (GST) was 41.4%, and the gene flow was relatively low (Nm = 0.7085) among X. italicum populations.
AMOVA results of X. italicum populations
| Source of variation | Degree of freedom | Sum of squares | Variance component | Percentage of variation (%) | P-value |
|---|---|---|---|---|---|
| Among populations | 9 | 87.5229 | 0.1404 | 22.72 | <0.001 |
| Within populations | 175 | 316.1974 | 0.4776 | 77.28 | <0.001 |
| Total | 184 | 403.7203 | 0.6180 | 100 | |
3.3. Genetic relationship among populations
The genetic distance (below diagonal) and genetic identity (above diagonal) between X. italicum populations is shown in Table 5, revealing that the genetic distance among populations ranged from 0.0704 to 0.5255 with an average of 0.2255. The highest genetic distance was between populations XA and SD, and the lowest genetic distance was between populations WLH and SD. An UPGMA dendrogram was obtained to estimate phylogenetic relationships among the 10 populations using genetic identity matrices. The 10 populations were separated into three groups (Figure 2). Pop1 to Pop4 formed Group I, Pop5 to Pop7 formed Group II, and the remaining populations (Pop8 to Pop10) formed Group III. A Mantel test of geographic distance and genetic distance was performed, and the results showed that there was no definite geographic trend in the distribution of the genetic differentiation (r = 0.1041, P = 0.3010).
Nei’s (1978) unbiased measures of genetic distance (below diagonal) and genetic identity (above diagonal) among X. italicum populations
| ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | **** | 0.8772 | 0.8703 | 0.8785 | 0.8288 | 0.7854 | 0.8840 | 0.7510 | 0.7287 | 0.7309 |
| 2 | 0.1311 | **** | 0.8408 | 0.8844 | 0.8682 | 0.7975 | 0.8458 | 0.7635 | 0.8042 | 0.7853 |
| 3 | 0.1389 | 0.1734 | **** | 0.9272 | 0.7768 | 0.6886 | 0.7881 | 0.6191 | 0.6510 | 0.5913 |
| 4 | 0.1296 | 0.1228 | 0.0756 | **** | 0.8673 | 0.8097 | 0.8461 | 0.7506 | 0.7593 | 0.7310 |
| 5 | 0.1877 | 0.1414 | 0.2525 | 0.1423 | **** | 0.8929 | 0.8926 | 0.7681 | 0.7860 | 0.7703 |
| 6 | 0.2416 | 0.2263 | 0.3730 | 0.2111 | 0.1132 | **** | 0.8191 | 0.7391 | 0.7282 | 0.7310 |
| 7 | 0.1696 | 0.1675 | 0.2382 | 0.1671 | 0.1136 | 0.1996 | **** | 0.8614 | 0.8397 | 0.8046 |
| 8 | 0.2864 | 0.2699 | 0.4796 | 0.2869 | 0.2638 | 0.3023 | 0.1492 | **** | 0.9315 | 0.9100 |
| 9 | 0.3165 | 0.2179 | 0.4293 | 0.2754 | 0.2408 | 0.3172 | 0.1747 | 0.0709 | **** | 0.9320 |
| 10 | 0.3135 | 0.2416 | 0.5255 | 0.3133 | 0.2610 | 0.3133 | 0.2174 | 0.0943 | 0.0704 | **** |
4. Discussion
4.1. Genetic diversity in X. italicum populations
The results of the POPGENE software analysis showed that the Nei’s genetic diversity (H) of X. italicum was 0.3673. Comparison with several alien species of the Compositae family by using ISSR markers revealed that the genetic diversity of X. italicum at the species level was significantly higher than that of plants in the same family, such as Galinsoga quadriradiata (HISSR = 0.0604) [30], Chromolaena odorata (HISSR = 0.0406) [31], and Parthenium hysterophorus (HISSR = 0.2887) [32].
Under natural conditions, the germination rate of X. italicum seeds is more than 90%, the flowering period is more than one month, and the average individual can form 6200 ripe fruits. Its pollen can be transmitted by wind, animals, or humans, its fruit quantity is large, and its fruit surface forms dense barbs [17]. X. italicum does not require strict soil and water conditions, and it is often found in habitats such as roadsides, wastelands, farmlands, and grasslands [15]. As a harmful weed with strong invasive ability, X. italicum has a wide distribution range and diverse habitat conditions, easily forms a dense single population, and can even secrete phytotoxic substances, resulting in the inability of other plants to survive around it [13]. These biological characteristics are suitable for its rapid diffusion and reproduction, and cross-pollination between different individuals provides an opportunity for gene recombination, thus improving genetic diversity. From the viewpoint of evolution, higher genetic diversity indicates strong potential fitness of an alien species to adapt to environmental changes and to expand its distribution range in new habitats [33], which may be an important factor causing the strong invasiveness of X. italicum.
4.2. Genetic differentiation among X. italicum populations
The total gene diversity (Ht) and gene diversity within X. italicum populations (Hs) were 0.3578 and 0.2098 respectively, which indicated that the genetic variation was mainly within populations. AMOVA results showed that there was a significant genetic variation among populations that accounted for 22.72% of the total variation, and the genetic variation within populations was 77.28%. This may have been the combined effect of gene flow and environmental selection pressure [34]. Compared to other invasive Compositae plants in China, such as Mikania micrantha (Gst = 36.49%) [35], Ambrosia artemisiifolia (Gst = 32.63%) [36], and Eupatorium catarium (Gst = 27.61%) [37], there was a very obvious genetic differentiation among X. italicum populations (Gst = 41.40%). It is generally believed that under the condition of gene flow (Nm) < 1, genetic drift will aggravate genetic differentiation [38]. The gene flow among X. italicum populations was 0.7085, which was consistent with the result of higher genetic differentiation among populations. This gene flow was regarded as a responsible factor for the genetic structure of the X. italicum population because introduction of new alleles into the population through gene flow is a very important source of genetic variation, which affects the genetic diversity of the population and produces a new combination of characteristics. In addition, the locations of these X. italicum populations are not only geographically far away from each other but also characterised by distinctive climates: the WLH, JZ, DWC, GHC, GT, and XA populations have a temperate continental monsoon climate; the LTS and SD populations have a temperate maritime climate; and the SHZ and YL populations have a typical temperate continental arid/semiarid climate [39]. Different climate conditions could cause different environmental selection pressures in these populations; therefore, X. italicum may rapidly accumulate favourable genetic variation to adapt to different climates and environments in a wide range of regions.
4.3. Genetic relationship and cluster analysis among X. italicum populations
The genetic variation of an invasive species can be used to determine its geographical origin and confirm whether it originates from single or multiple populations [40]. The quarantine data of X. italicum from various customs in China showed they came from three different countries [41, 42, 43]. Combined with the results of cluster analysis, it was inferred that X. italicum might have come from three different origins. Mantel test results showed that there was no significant correlation between genetic and geographical distances among X. italicum populations (r = 0.1041, P = 0.3010). The GT population was geographically distant from the GHC and DWC populations, but their genetic distance was relatively close and clustered into one group; the XA population was geographically near to the WLH and SD populations, but their genetic distance was high and showed no aggregation. The long-distance spread of X. italicum is accomplished by anthropogenic activities, such as transport of goods and trade, but its short-distance spread is accomplished by animals, wind, and water [44]. Therefore, it was speculated that the pathways of X. italicum invasion may be artificial and natural diffusions. For example, the YL, SHZ, XA, WLH, JZ, LTS, and SD populations were far apart, and thus might be diffused by human factors. However, the GT, GHC, and DWC populations are relatively close to each other, and thus may be formed through natural diffusion of an invasive population that produced a large number of seeds.
Moreover, most of the genetic variation of invasive plants usually exists among populations [45], whereas that of X. italicum principally exists within populations. One possible explanation for this finding was that disturbance by anthropogenic activities (such as transportation of humans, seedlings, livestock, and cereals) affected the distribution and gene flow of X. italicum populations, which led to genetic differentiation and changes in its population genetic structure. The seeds of X. italicum have barbed thorns on their surface, making its seeds easy to be carried by humans or animals. This leads to the occurrence of multiple introduction events, thus improving the genetic communication between X. italicum populations. Another explanation was that two species of the Xanthium genus, X. sibiricum and X. spinosum, have the same domain distribution and hybridisation with X. italicum [17], which likely produced hybrid populations, altered their genetic differentiation, and promoted its successful invasion [46]. As a result, the genetic diversity of multiple introductions and genetic recombination endowed the alien species with higher evolution potential and invasiveness, allowing it to occupy wider distribution areas in the course of introduction and to generate greater harm.
Alien species that experienced genetic bottleneck eventually develop into successful invasive species. This is known as the “genetic paradox”. Multiple introductions can make up for the decrease in genetic diversity caused by a founder effect or bottleneck effect to a great extent. The effects of multiple introductions and founder effect on genetic diversity are antagonistic, but they could occur in different invasion events of the same alien species [47]. Our results showed that X. italicum populations had very high genetic diversity, with the genetic variation mainly occurring within populations, and the pattern of its existing genetic structure was related to its biological characteristics and human disturbance. It is generally believed that two possibilities can lead to higher genetic diversity in invasive plant populations: (i) existing weed populations may be established by individuals from multiple different sources, allowing them to maintain higher levels of genetic diversity; (ii) other weed populations may be established by a small number of founder individuals, but in the stage of plant population development, pollen flow and seedling regeneration can introduce additional genetic diversity, which leads to higher genetic diversity of the populations.
ISSR marker technology was an effective tool for studying gene polymorphism of alien species. However, it mainly marks the dominant gene, which does not distinguish heterozygote from homozygote, so that the genetic information is limited. The use of more co-dominant markers and sequence data from cpDNA or nrDNA (ITS) to study genetic diversity and gene flow, would be advantageous and would allow us to better understand the invasive mechanisms of this plant. In conclusion, our results show the high genetic diversity of X. italicum, which may help explain its invasion success in China. They support the first hypothesis that Chinese X. italicum populations probably dispersed from three different locations, according to the genetic and customs data of this weed in China. However, more accurate genetic information about this alien plant is required. Thus, a comparative study of genetic diversity between X. italicum populations in their original habitats and in invaded areas is a certainly interesting and important topic for future research, as it might be relevant to determine the origin of this plant as well as to decrease and eventually stop its invasion in China.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (project number: 31360047).




 CC-BY 4.0
CC-BY 4.0