1 Introduction
Sheep scrapie, soon recognised as a transmissible spongiform encephalopathy (TSE), or prion disease 〚1〛, is the most widespread animal TSE in the field where it can spread following an infectious mode 〚2〛. Despite intensive studies performed during the last decades on sheep scrapie, a number of crucially important questions, such as the natural mode of sheep to sheep transmission, and the diversity and possible evolution of the strains, which circulate in the field, remain largely unanswered. The consecutive emergence of bovine spongiform encephalopathy (BSE) epidemics in cattle and of the variant Creutzfeldt-Jakob (vCJD) in humans have greatly stimulated the research efforts on sheep scrapie, notably in view of the potential sheep origin of BSE and of the serious consequences that an accidental transmission of the bovine agent to sheep would entail for public health 〚3〛. The present review deals with newly introduced models, mice and cell lines genetically engineered for ovine PrP expression, that have been developed in our laboratory with the aim of increasing the possibilities of investigations on the sheep infectious agent.
2 Transgenic mice as an alternative animal model to study the natural sheep scrapie agent
Most of the experimental work accomplished up to now on the sheep scrapie agent has been based on its inoculation to either the natural host, or standard laboratory rodent animals, mostly mice and hamster. Such studies have enabled impressive advances in understanding TSE diseases in many respects. In particular, they have revealed that susceptibility to the disease is the result of an intimate and complex interplay between the host and the causal agent. The observed complexity is partly accounted for by the diversity not only of the gene encoding the PrP protein but also of the agent itself, which is comprised of a number of strains exhibiting distinctive biological properties, and this, irrespective of the host genotype 〚4–9〛. Passaging of scrapie isolates by inoculation into a panel of congenic mice, where characteristics of incubation time and brain vacuolation areas give distinct patterns, is currently the most appropriate way to investigate strains 〚6, 10〛. Such a procedure, however, suffers from two main drawbacks as far as elucidating the extent and basis of the natural diversity of the sheep TSE agent is concerned. Firstly, it implies propagation of the agent on a mouse, not a sheep PrP background. Secondly, even after intracerebral inoculation, the disease commonly requires 1 or 2 years to develop, and a number of isolates do not transmit easily to mice.
As demonstrated by former studies on other TSE agents, including bovine and human ones, mouse transgenesis provides unique opportunities in these regards 〚11–16〛. First of all, this approach makes it possible to analyse the behaviour of a TSE agent on a PrP background homologous to that of the donor species. In addition, transgenesis enables to overexpress a relevant PrP gene, thus contributing to increase the efficiency of the transmission of the disease to the recipient mice compared to non-transgenic mice. These observations have led to the proposal that expression of a PrP gene homologous to that of the prion donor species may be sufficient to abrogate the interspecific barrier 〚11〛, even though an enhanced transmission is not the universal outcome 〚17〛.
In an effort to develop an improved model for transmission of sheep scrapie, we have expanded a series of transgenic mice expressing an ovine PrP gene (tgOv), and examined their susceptibility to sheep isolates in primary transmission 〚18〛. All these lines express the so-called VRQ allele of PrP (136Val-154Arg-171Gln), which has been found to be associated in sheep with both an extremely high susceptibility to the disease and a relatively shorter incubation time in independent studies 〚7–9, 19〛. Each of them expressed this PrPVRQ transgene on an ablated mouse PrP0/0 background 〚20〛, since co-expression of the endogenous PrP gene was reported to generate an interfering effect 〚13, 21〛. These tgOv differed, however, according to the level of PrP expression in their brain, and also the type of the transgene construct. The tg1 construct was derived from the phgPrP half-genomic vector, already validated for PrP transgenesis 〚14〛, by just replacing the mouse open reading frame by the ovine one. The tg2 construct was derived from the former by substituting the human cytomegalovirus promoter to the PrP murine promoter sequences. The tg3 construct corresponds to a 125-kb sheep DNA BAC insert encompassing the whole Prnp transcription unit. In each case, the transgene was introgressed for at least three generations on the PrP0/0 knockout mouse line. All of the 10 tgOv lines obtained were tested for their susceptibility at the heterozygous state, by using two isolates issued from scrapie-affected, VRQ homozygous sheep. Two high expressing tg3 lines, tg301 and tg338, were tested also at a transgene homozygous state.
From the main results, illustrated in figure 1, the following conclusions could be drawn:
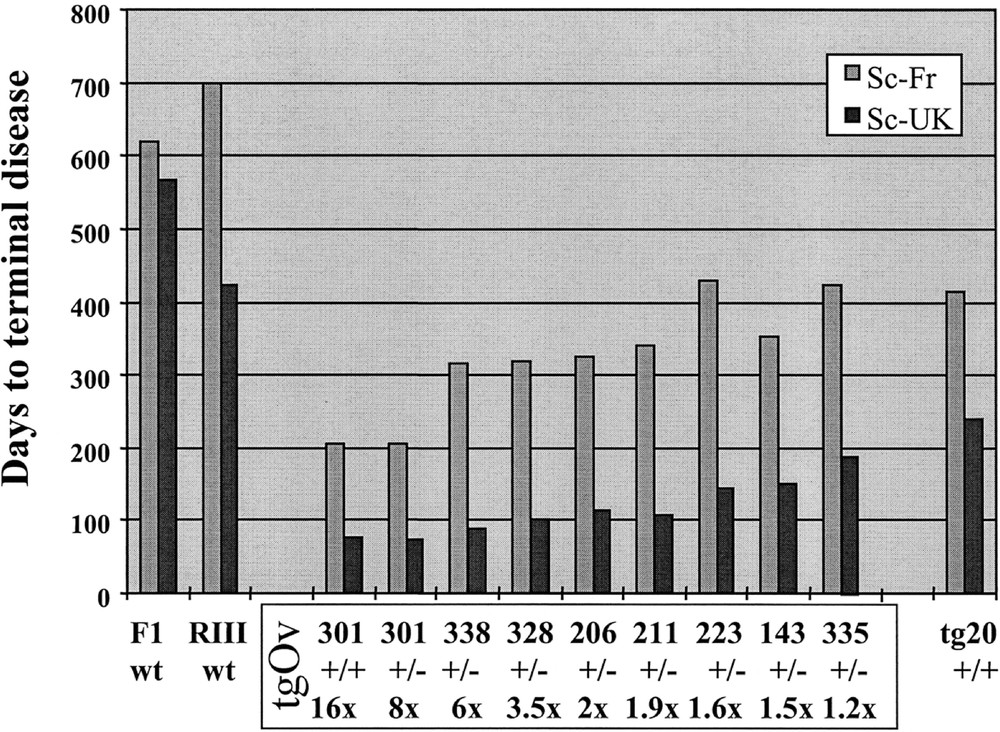
Primary transmission of natural scrapie to ovine PrPVRQ transgenic mice. The survival time of 8 mouse transgenic lines (tgOv) expressing ovine PrP at various levels from 3 different constructs is shown. Each line is designated by a number of which the first digit refers to the construct (tg1, 2 or 3; see text). Their respective brain PrP protein level of expression is expressed in –fold that in sheep brain. The survival time of wild-type mice (F1-C57BL/6 × 129/sv and RIII) and of transgenic mice overexpresssing mouse PrP (tg20; 〚14〛) is shown for comparison. Each animal was inoculated intracerebrally with 10 % heated homogenate of brain material derived from either a French (Sc-Fr) or a British (Sc-UK) field scrapie case. The observed survival time differed markedly between the 2 isolates, both originating from a sheep homozygous for the VRQ allele (e.g. 73 + 3 and 205 + 12 days for Sc-UK and Sc-Fr isolates, respectively, in the tg301 line heterozygous (+/–) for the transgene).
- • as a general rule, a 100 % attack rate was observed whatever the tgOv line and the isolate. All mice showed acute neurological disorders typical of TSE, and accumulated abnormal PrP 〚22〛 in their brain, as revealed by resistance to PK digestion (PrPres) and western blotting. The disease failed to be transmitted in only one case, the low PrP expressing tg116 line (< 0.4-fold that in sheep brain) after challenge with the French isolate, although a few animals accumulated brain PrP-res before they died.
- • for both isolates, the survival time was substantially reduced when compared to those of the wild-type, RIII and (C57Bl/6 × 129/sv)F1 mice.
- • the survival time appeared to be correlated essentially with the brain PrP content, which varied from 16-fold to < 0.4-fold in these experiments. No obvious effect of the type of construct could be noted, at least after intracerebral inoculation, the only route studied so far.
- • in the mouse lines expressing ovine PrP at the highest level (≥ 8 ×), the incubation duration was about 2 and 7 months for the UK and the French isolate, respectively, i.e. one year or more briefer than in the conventional mice.
- • the tga20 mice 〚14〛, overexpressing a mouse PrP transgene at a level equivalent to tg301+/– mice on a similar genetic background, were found to be markedly less susceptible than the latter, and as susceptible as the tgOv lines expressing the transgene at a nearly physiological level.
While providing clear evidence that transgenic expression of ovine PrP may confer increased susceptibility to natural sheep scrapie on mice, this study leaves open a number of issues that need to be addressed in future investigations.
- • “Was expression of a VRQ allele instrumental for an improved transmission compared to non-transgenic, conventional mice?” The present data strongly argue that this allele behaved as a better substrate for sheep prion replication than did the mouse Prnpa allele, at least for the two analysed isolates. Another high susceptibility allele, ARQ (Ala instead of Val), also regarded as the ancestor ovine Prnp gene, is much more widespread within the sheep population 〚7, 8, 23〛. As a consequence, the majority of the scrapie-affected animals bears this allele, and several groups have favoured it for expression as a transgene. Several years ago, transgenic lines were created using a sheep cosmid harbouring the ARQ allele 〚24〛. Later on, it was quoted in a review that disease transmission in those mice was not improved when compared to conventional mice 〚25〛. The transgene, however, was not expressed on a PrP0/0 ablated background (see above). That this point may be of concern is supported by results from ongoing experiments on tgVRQ mice, showing that co-expression of the mouse PrP results in a protracted survival time after infection with the fast (see figure 1), UK isolate. More recently, two other groups have derived mice expressing the ARQ allele through two different constructs, both in the absence of endogenous PrP, and reported an enhanced transmission compared to conventional, C57BL/6 mice 〚26, 27〛. However, whether such tgARQ mice would exhibit a higher susceptibility than transgenic mice overexpressing mouse PrP at the same level, as appeared to be the case for the tgVRQ mice, remains to be established. A comparison of the various tgOv mice now available should make it possible in the near future to clarify which of the VRQ or ARQ alleles would impart a more favourable context for replication of most scrapie strains, even though formal assessment of this point would require a gene replacement approach.
- • “Did the expression of an ovine PrP gene erase the sheep to mouse barrier?” Pioneering studies dealing with characterising the sheep scrapie agent have shed light on its amazing diversity 〚4, 6〛, and led to the proposal that it may embrace as many as fourteen phenotypically distinct strains 〚28〛. One should bear in mind, however, that this notion relies on transmissions from sheep to mouse, also involving in a number of cases intermediate hosts such as a goat or a mouse with another Prnp genotype. As species barrier crossing may enhance prion diversity 〚6, 29, 30〛, or reduce it by failure to transmit 〚10, 31〛, a precise idea of the natural scrapie strain diversity has yet to be gained. Whether the sheep agent would be subjected to a lower selection pressure when transmitted to tgOv mice instead of conventional mice does matter in this regard. One possible criterion to address this question is the evolution of the incubation period on further passaging in the recipient mice. In this study, serial passages performed on one of the lines (tg211) did not lead to significant change in the survival time for either isolate 〚18〛. This is in sharp contrast with the marked reduction commonly observed on second passage in conventional mice, and suggests that transmission of the sheep agent to tgOv mice required little or no adaptation to its new host. This issue needs to be further addressed, however, since present knowledge does not allow excluding completely a phenotypical shifting of the sheep agent when passed on tgOv mice.
- • “TgOv-VRQ mice: an improved tool for field scrapie strain typing?” As pointed out above, tgOv mice could enable a more faithful assessment of the nature and extent of the variation among the field scrapie strains. However, it has yet to be determined: i) how the mice will respond to an enlarged panel of isolates; ii) to what extent the PrP genotype of the donor sheep will influence the efficiency of transmission: indeed, this study was focused on isolates from a homozygous VRQ individual, thus resulting solely in homotypic transmissions. Documenting both of these points is a goal of ongoing studies. To date, a few more scrapie isolates, including isolates issued from ARQ donor sheep, have been very efficiently transmitted to these mice. However, owing to the existing evidence that certain sheep or mouse-adapted strains exhibit striking differences in terms of allele preference 〚5, 7, 31〛, it seems unlikely to us that tgOv-VRQ mice would represent a universal receiver for field scrapie strains. In any event, it seems reasonable to predict that substituting congenic, wild-type mice with transgenic mouse lines, each overexpressing a specific PrP allele, will offer a time-saving strategy for strain typing. As a matter of fact, transmission to tgOv mice enabled a clear differentiation of the two isolates studied within a much shorter period of time than in conventional mice.
- • “Would tgOv mice provide not only a faster but also a more sensitive infectivity assay of the sheep infectious agent?” Apparently, heightening mouse PrP expression does not confer an enhanced sensitivity on mice in terms of detection limit 〚see 14〛. Published data regarding the effect of the expression of a heterologous transgene are still lacking. Titration of a few sheep isolates in tgOv mice is currently in progress. Rigorous assessment of this point, however, would require parallel endpoint titrations in mice expressing various levels of the transgene and wild-type mice, or in transgenic mice and the natural host. Such experiments are underway in the case of bovine PrP transgenic mice 〚32〛, and could be informative in this respect. It remains that determining prion infectivity through bioassay in transgenic mice would provide only limited improvement from a practical and ethical point of view. Accordingly, increased efforts should be made in an attempt to develop cell culture-based infectivity assays as an alternative to in vivo assays.
3 Propagation of natural sheep scrapie infectious agent in cell culture: toward new TSE cellular models
Despite repeated attempts throughout the last three decades 〚33〛, only a few cell models sustaining prion propagation in culture have been developed successfully to date. Among them, PC12 rat pheochromocytoma cells 〚34〛, N2a mouse neuroblastoma cells 〚35〛, and more recently, GT1 hypothalamic neurons 〚36〛 have been the most commonly used infectable cell lines. Such systems, as well as the persistently infected SMB murine cell line 〚37〛, allow both active replication of the infectious agent and accumulation of abnormal, PK-resistant PrP in readily detectable amounts. They have brought valuable insights into the study of TSE infection at a cellular level in several respects, notably the subcellular distribution of the abnormal PrP, and the key role of specific PrP amino acid residues in the PrP to PrPsc conversion process (for a review, see 〚38〛 or Lehmann et al., this issue). A common feature of the above models, however, is to support the multiplication of laboratory, rodent TSE strains. Indeed, relevant cell systems for strains from naturally occurring TSE diseases such as sheep scrapie, bovine spongiform encephalopathy, and Creutzfeldt-Jakob disease are lacking so far.
In an attempt to establish new permissive cell systems potentially enabling an in depth study of TSE causing agents without prior adaptation to the rodent, various strategies have been explored in our laboratory. While the sheep agent was chosen as a primary target, part of the work was intentionally focused on non ovine cells, assuming that any conclusive approach could tentatively be exploited in the aim of deriving further cell systems permissive to prions affecting other species. It should immediately be added that, not surprisingly, pursuing this appealing but somewhat ingenuous task was not exempt of disappointments. In figure 2 are presented three approaches, which illustrate our exploratory work.
Development of new TSE cell systems. Three different strategies that have been explored in order to establish cell lines able to replicate ovine sheep TSE agent are illustrated.
- • “Cell lines immortalised from sheep nervous tissue.” The rationale of this approach is based on the fact that most, if not all – PC12 cells are from rat – the existing systems permissive to mouse prions are based on neural cells derived from the homologous species. Rather unfortunately, such cell lines were lacking in the case of sheep. We have thus undertaken to establish such material by using protocols originally developed for rodent 〚39〛. Primary cultures were established from the brain of 2 month-old sheep embryos of the relevant genotype, and were transfected with the SV40 large T protein gene 〚40〛. Two of the so-obtained cell lines, A15 and 4A6, displayed features of astrocytes 〚41〛. They were studied in more detail since there is indirect evidence that such cells may support prion replication in vivo 〚42〛. Cultures were transfected so as to heighten the ovine PrP expression level 〚43〛. One 4A6 line-derived cell clone with a 4-fold increased PrP expression could be successfully infected with the fast scrapie isolate. Indeed, cell extracts from 8-fold passaged cultures killed all the inoculated tgOv331 mice in about 3 months (unpublished data). However, PrP-res accumulation was never observed in such cultures, consistent with the relatively low level infectivity compared to the original inoculum. Immortalisation of cells derived from adult sheep infected brain was also attempted. None of the twenty cell clones obtained in this way accumulated PrP-res at a detectable level.
- • “Ovine PrP genetically engineered non-ovine cell lines.” This approach, although somewhat provocative in view of what was stated just above, was not so irrational as it might appear at first. After all, transgenic mice, including ‘ovinised’ mice, are able to replicate prion from foreign species (see above), and we speculated that the species barrier could be crossed ex vivo as well, at least with a bit of luck and a grain of salt. The salt consisted in i) selecting among a set of widely used cell lines those in which endogenous PrP was naturally expressed at a low or undetectable level; and ii) isolating transfected clones that exhibited a high level of ovine PrP expression. Their tissue origin was not taken into account – and here perhaps lies the luck. It was also decided to use a vector (Tet on) allowing a regulable expression of the transgene 〚44〛, in particular so as to facilitate assessing the infected status in inoculated cultures. It turned out that the first cell clones to meet the above-mentioned criteria were derived from the rabbit kidney, epithelial cell line RK13, leading to the TSE cell system named Rov 〚45〛.
All of the Rov cell clones selected from independent transfection experiments, and then tested for their permissiveness were found to be readily and consistently infectable by the sheep agent. Unlike scrapie-inoculated astrocyte-like and non-induced Rov cell cultures, PrP-induced, infected Rov cell cultures accumulated PK-resistant PrPres at a substantial level, which compares with that found in a scrapie sheep brain. De novo produced PrPres could be detected from the first subpassage post-infection, and reached a plateau after 10–15 subpassages (the cultures are subjected to a weekly splitting). Chronically infected cultures were propagated for many passages in the absence of any noticeable cytophatic effect. In such cultures, the proportion of PrPres positive cells averaged 50 percent, as determined by immunochemistry examination of pelleted, paraffin-embedded material. Such a relatively high rate of infection may explain why the latter propagates steadily without the need for subcloning, contrary to other TSE-permissive cell systems 〚46〛. Extracts prepared from a few million cells of infected cultures that were subpassaged 8-fold or more were able to induce classical TSE neurological disorders, leading to the death of all of the recipient tgOv mice within a period of time similar to that observed with a 10 % homogenate from the original sheep brain. Bioassay of extracts from inoculated, 8-fold passaged parental RK13 cell cultures repeatedly failed to evidence any infectivity. Altogether, these results demonstrated that expression of ovine PrP otherwise refractory, rabbit cells conferred on them the capacity to actively propagate a not only infectious, but also true scrapie-engendering agent.
– “Cell lines derived from ovine PrP transgenic mice.” Deriving stable cell lines from mice transgenic for SV40 large T antigen, an immortalising protein, has proved to be an efficient strategy in order to establish cells from a specified tissue origin in culture 〚47〛. Thus it was tempting to apply such an approach to tgOv mice to obtain ovine PrP-expressing cell lines from different tissues that represent natural targets for TSE agents. To do so, mice expressing SV40 antigen on a PrP0/0 background were established, then crossed with tg301 tgOv mice. Several stable cell clones were established from the nervous system, and are currently being tested for their permissiveness to the sheep agent. Although somewhat laborious, this approach looks promising. Indeed, two independent clones were found to accumulate PrPres in substantial amounts upon infection, and to propagate the sheep agent at infectivity levels similar to that of Rov cells, as judged from bioassays in tgOv mice. The precise nature of these cells, which derive from peripheral nervous tissue, is under investigation.
Each of the three above approaches allowed us to establish new TSE cell systems, in which a natural, non rodent-adapted TSE agent can be multiplied and studied ex vivo. In two of them, the efficiency of replication appears to be comparable to that in the original brain tissue used as inoculum, according to both the level of abnormal PrP accumulation and infectivity. From these observations, we would like to emphasise the following points.
- • Several unique features make the Rov cell system an attractive model in the aim of investigating essential aspects of TSE infection at the cellular level. Firstly, it should give access to reverse genetics, through the selection of transfected clones expressing various alleles or genetically modified PrP genes, followed by the examination of their respective ability to restore susceptibility to the infection. Unlike the N2a cell system to which similar approaches have been applied 〚48, 49〛, it will be feasible here to assess any effect resulting from a modification in the PrP sequence not only on the PrP conversion process but also on the infectious agent replication, and this in the absence of the wild-type PrP. Thus, cell clones expressing various ovine PrP alleles, including the ‘resistance’ ARR allele 〚7–9〛, are currently being derived in our laboratory in order to establish whether the striking effect of the PrP polymorphism on the vulnerability of sheep to scrapie can be mimicked ex vivo. Secondly, the availability of cell clones combining infectability and regulable PrP protein expression might facilitate further investigations dealing with specific aspects of the infectious cycle of a prion: is PrP needed during the early phase of the infection, and what are the effects of turning off PrP once infection is established?
- • A common feature of two of the above strategies is that propagation of sheep agent was achieved in non-ovine cells, either from rabbit or mouse. Hence, they could potentially be extended to TSE affecting other species, including the human and bovine ones, a possibility which is currently under investigation. It should be kept in mind, however, that there is some evidence that host factors other than PrP might control interspecies transmission, and this possibly in a strain-dependent manner 〚13, 17〛. It is thus reasonable to expect that a situation similar to that in transgenic mice could also be observed at the cell level, i.e. engineering heterologous cells with a cognate PrP gene would not always suffice to confer on them an enhanced permissiveness to the relevant TSE agent. An ex vivo approach might facilitate investigations about whether the species barrier is also mediated by species- or strain-specific interactions between PrP and cellular factors, as opposed to prion protein-protein interactions 〚49, 50〛.
- • Further studies will be needed to assess whether the scrapie infectious agent propagated in either mouse or rabbit cells fully retains its original strain phenotype. Proteinase K-digested PrP (PrP-res) of brain tissue from tgOv mice inoculated with either the original sheep brain homogenate or infected cell extracts showed indistinguishable banding patterns in immunoblot. Worthy of note, however, the patterns generated from tgOv brain, sheep brain and cell culture infected materials differed strikingly from each other. This and earlier observations 〚51, 52〛 strengthen the view that the PrPres profile is not an intangible strain marker, but may also depend on the environment in which the abnormal PrP is produced.
- • The finding that epithelial cells can sustain active prion replication in culture raises several stimulating issues. Firstly, the nervous system is developmentally derived from an epithelium, and both neurons and epithelial cells are highly polarised. Furthermore, similar mechanisms have been proposed to act for the targeted sorting of certain proteins common to both types of cells 〚53, 54〛. Secondly, the spreading of prions within an infected organism appears to involve markedly distinct types of cell actors, of which it is reasonable to think that the inventory as well as the elucidation of the respective contribution is far from complete. Epithelial cells are present in several organs, placenta 〚55〛, digestive tract 〚56〛 and skin 〚57〛 that are actually or potentially involved in TSE pathogenesis. Their possible involvement in prion docking, transport or replication would deserve closer investigation. In particular, the role of the cells lining the lumen of the digestive tract needs to be further examined, since PrP is expressed at a substantial level 〚58〛 in this epithelium that has to be crossed by the infectious agent after oral contamination. More generally, an ex vivo approach could contribute to identifying cell types that are presently not regarded as targets for prions.
4 Concluding remarks
Mice endowed with an increased susceptibility to the sheep agent provide another example of the unique contribution of transgenesis to TSE research. There is little doubt that such tgOv mice will bring new opportunities for characterising scrapie strains, measuring infectivity, and more generally analysing the mechanisms underlying the pathogenesis of the disease. An unanticipated outcome of this work was the availability of a new TSE model with one of the shortest incubation times – two months in primary transmission – of any such model. On the other hand, the introduction of newly derived cell lines that enable an efficient propagation of the sheep agent in culture should provide an incentive for exploring various strategies toward the development of more diversified and powerful TSE cell systems. Finally, the research on prion diseases should benefit from the combined and reciprocally enforced use of such in vivo and ex vivo models.
Acknowledgements
We are grateful to many colleagues for providing help, material or support: O. Andreoletti (ENVT-INRA Toulouse), A. Buschmann (FCVDA Tübingen), B. Cayron (INRA Jouy; animal care), J. Costa Da Silva (GBC INRA Jouy), E. Cribiu (GBC INRA Jouy), M. Dawson (CVL Addelstone), J.P. Deslys (CEA Fontenay), J.M. Elsen (INRA Saga Toulouse; Sc-Fr isolate), M. Groschup (FCVDA Tübingen), C. La Bonnardière (VIM INRA Jouy), T.L. Lai (VIM INRA Jouy), S. Lehmann (IGH-CNRS Montpellier), M.F. Madelaine (VIM INRA Jouy), D. Paulin (Paris VI University; SV40 T antigen plasmid and transgenic mice), S. Rakotobe (VIM INRA Jouy), P. Sarradin (INRA Tours; NPU Edinburgh RIII mice), M.G. Stinnakre (GBC INRA Jouy), D. Vaiman (GBC INRA Jouy; BAC library), and C. Weissmann (phgPrP vector, PrP°/° and tg20 mice). This work was supported by grants from the European Community (FAIR 5CT-973304 and BIOTECH PL-976064) and from the French government (Cellule de Coordination Interorganisme sur les EST).



