1 Yeast cellular morphotypes
The smallest replicative unit of a eukaryotic cell is constituted of the nucleus, which however relies on the cytoplasm for basic functionalities required for replication. In turn, the smallest functional entity that makes up an organism is a cell. From this duality – cells and nuclei – results various basic morphotypes (Fig. 1).
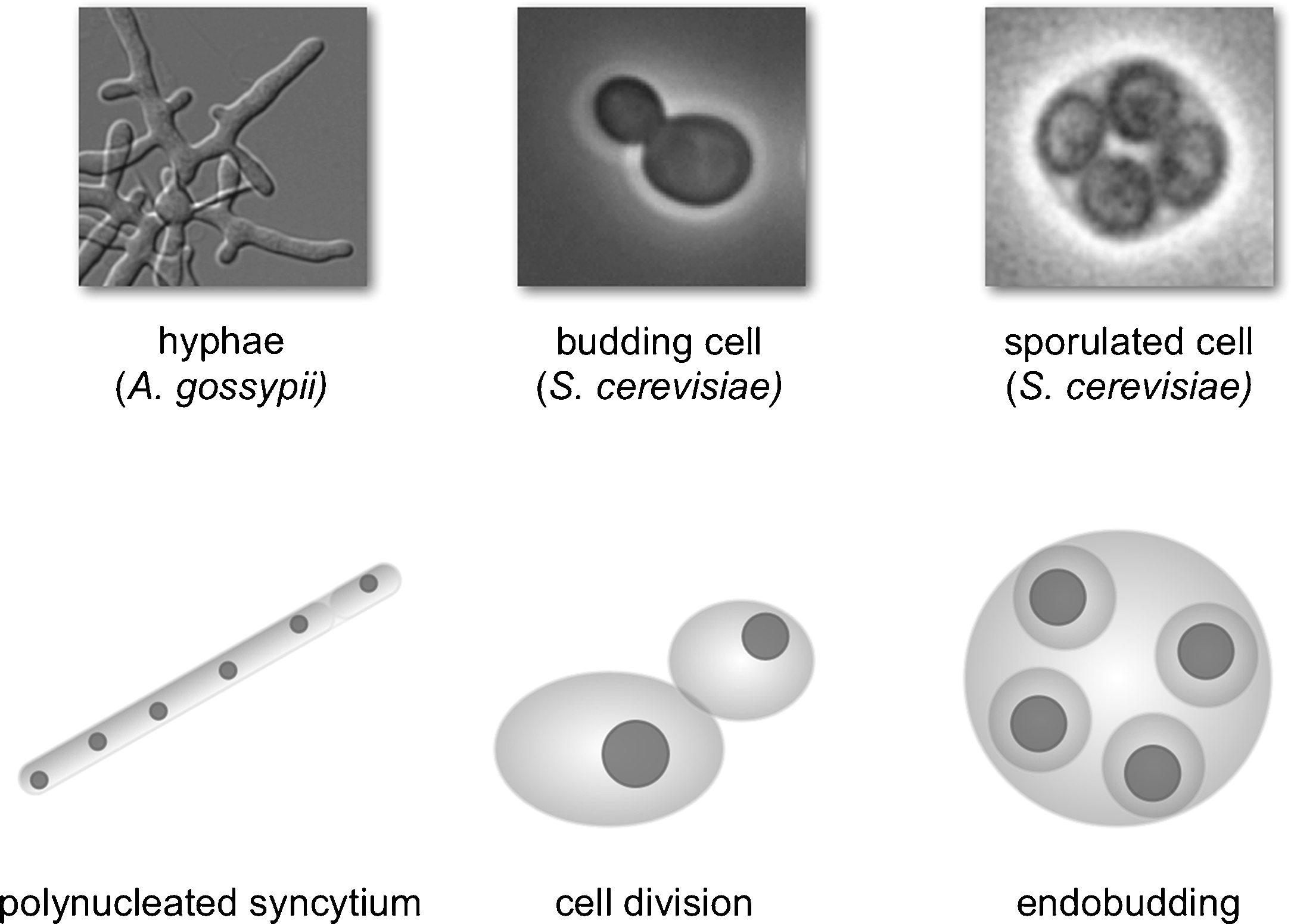
Basic cellular morphotypes. Hyphae of the yeast species A. gossypii are shown as an example of a polynucleated syncytium (left panels). Vegetatively yeast cell of round or ellipsoid shape divide almost exclusively via bud formation adjacent to a mother cell (middle). Spores are assembled inside the mother cell by endobudding.
The first morphotype is the syncytium, defined as one cytoplasm containing many nuclei. This morphotype is not very frequently observed in yeasts. Yeast species able to form hyphal syncytia (true septate hyphae), also called filaments, are for example represented by the strictly hyphal species Ashbya gossypii from the genus Eremothecium. In hyphae of A. gossypii, the replicative cycle of individual nuclei is truly uncoupled from cell growth and the processes in the cytoplasm. In fact, each nucleus is able to follow its own replicative cycle, irrespective of the replicative stage of neighboring nuclei [1]. This nuclear autonomy raises a number of interesting questions in terms of the molecular control mechanisms of nuclear division.
The second morphotype is the ‘normal’ one nucleus-one cell arrangement, where nuclear division and cell division are tightly coupled via the cell cycle machinery. This coupling is evident during purely vegetative replication cycles of yeast cells. For many species this normal cell division occurs also in the first cell division after mating and zygote formation.
In other species mating may be immediately followed by meiosis and spore formation. This leads us to the third relevant morphotype, which is cell division by endobudding, a process comparable to endodyodeny or endopolygeny in many protists such as Toxoplasma gondii. Here, cell replication takes place by the assembly of daughter cells inside the mother cells from nuclei that have been generated in one to several preceding nuclear divisions. This type of cell division involves a break of topology whereby each nucleus organizes its separation from the common cytoplasm by assembling a new plasma membrane that is discontinuous from the syncytial plasma membrane. In the yeasts, endobudding appears to occur only when linked to spore formation, which involves the concomitant assembly of a spore wall. Although this form of cell division is prevalent for the yeasts, there appears to be no yeast species that uses consecutive cycles of endobudding as a mode of reproduction, as it is for example the case for the pathogenic form of the ascomycete fungus Coccidioides immitis.
2 Cell division by budding
Yeasts are classified as fungi whose primary morphological form is a unicellular cell form that divides by budding or fission. The vegetative cells of many yeast species that belong to the family of the Saccharomycetaceae or nearby genera (notably those sequenced by the Génolevures consortium [2,3]) are of similar ellipsoid or spherical shapes a few micrometer in diameter and divide by budding. These cells can be easily detected by microscopy because of the large refractive index difference between the cellular exterior and the cellular interior. Mature cells, termed ‘mothers’, form their ‘daughters’ via the formation of buds usually at one of the opposite poles of the spherical cells. The buds grow to reach a diameter that is typically smaller than the diameter of the mother (in Saccharomyces cerevisiae roughly 2/3 of the mother cell) before the physical disconnection of their cytoplasms in a process called cytokinesis.
This method of cell division, whereby a daughter cell forms adjacent to the mother cell that itself does not change in shape during the reproduction process, is probably the major unifying characteristic of yeast species. It constitutes an extreme version of asymmetric cell division, and as such contrasts the more ‘classical’ cell division that involves partitioning of the original cell into two equal cells found in many other organisms including bacteria, many mammalian cells and also the other important yeast model, the fission yeast Schizosaccharomyces pombe. The need to segregate cellular content from the mother compartment into the bud in every cell division made S. cerevisiae also an important model for studying asymmetric cell division. It is now utterly clear that yeasts have evolved mechanisms that allow a carefully monitoring the cellular content that should – or should not – be segregated into the bud [4,5].
3 Morphology of dimorphic transitions
Many, but not all yeast species undergo a major morphogenic transition: the dimorphic transition to pseudohyphal or hyphal/filamentous growth. Dimorphism denotes the phenotypic switching between two cell types. The transition from unicellular growth to pseudohyphae or true septate hyphae (filaments) is typically triggered by adverse environmental conditions. This distinguishes yeasts from many yeast-like fungi, mainly basidomycetes, which are primarily filamentous and respond to environmental stress by the generation of unicellular yeast-like forms. Characteristic features of the pseudohyphal growth are chains of markedly elongated cells that retain their unicellular character and that form new cells by a budding process (hence the name ‘pseudo’-hyphae). It has been demonstrated that the dimorphic transition can be gradual and able to generate a broad range of cell shapes, each specific of a response to a specific environmental parameter setting [6]. In S. cerevisiae the pseudohyphal transition involves a switch from a bipolar to a unipolar budding pattern. This generates, together with the maintenance of weak cell-cell contacts after cytokinesis, an asymmetric polarized cell colony that “permits otherwise sessile cells to forage for nutrients and substrate at a distance from their initial colonization site” [7].
Cell shape generation in pseudohyphae can be understood as a variation of the mechanisms that regulate the timing and polarization of vegetative cell growth and division. In contrast, true hyphae, which consist of syncytia with many nuclei in one cytoplasm, require an uncoupling of polarized cell growth from cell division (septum formation) and from nuclear divisions. One advantage of hyphae is related to the fact that the cytoplasm is disproportionally larger to the growth zone of the cell membrane as compared to the situation in a budding cell. Hyphae thus have an enormous capacity to drive the growth of the hyphae tips. For example, the hyphae of A. gossypii exhibit tip growth speeds in the range of 200 μm per hour [8], whereas budding cells grow 3–4 μm per hour.
4 Spore formation
The morphogenic transition that leads to a sporogenic form starts with either of the two vegetative dimorphic variants, budding cells or pseudohyphae/hyphae. Spore formation is usually preceded by meiosis, and thus leads to the formation of meiospores. The well-established order of events in the yeasts, namely the occurrence of meiosis before sporulation and subsequent germination is not necessarily the case in other fungal phyla. Spore formation in Microbotryum for example precedes the meiotic cell division, which only takes place during germination of the spore [9]. In other species, spores are often formed also without meiosis, in which case they are called mitospores (see below).
Spore formation occurs by endobudding, whereby multiple nuclei that are present in a single cytoplasm at the end of meiosis become separated from the common cytoplasm. The process is analogous to the transition from a multinucleated syncytium to a one-nucleus-one-cell state by means of encapsulation of the contained nuclei into individual compartments.
Spore formation after the meiotic division in Saccharomycetaceae species leads to tetrad formation in many species of different genera. In others, e.g. Kluyveromyces polysporus, subsequent mitoses increase the number of nuclei, which then become encapsulated into asci containing up to several dozens of spores [10]. Purely mitotic spore formation occurs for example in A. gossypii, where nuclei acquire spore walls inside segments of the hyphae, so-called sporangia [11]. A common theme for the regulation of spore formation in yeasts is that membrane formation of the cell/spore is not delegated to the cell polarity machinery. Instead, the spindle pole bodies (SPBs), the centrosome of yeasts, takes over the function of assembling the new spore membrane [12–14]. In yeast the nuclear membrane remains intact during nuclear divisions and the SPB is embedded in the nuclear envelope. This constellation permits the precise specification of the number of spore membranes that are required to engulf faithfully the products of the nuclear divisions.
5 The interplay between cell morphogenesis and life cycle
The cellular decisions to undergo morphogenic transitions that are related to the sexual life cycle, namely mating and meiosis, require an integration of external factors (available nutrients in the surrounding medium) with the internal status (e.g. haploid versus diploid, amount of available stored nutrients). Cells commit to these transitions in the G1 phase of the cell cycle, as only nuclei at the G1 stage can undergo the necessary transformations, e.g. in mating or meiosis (Fig. 2).
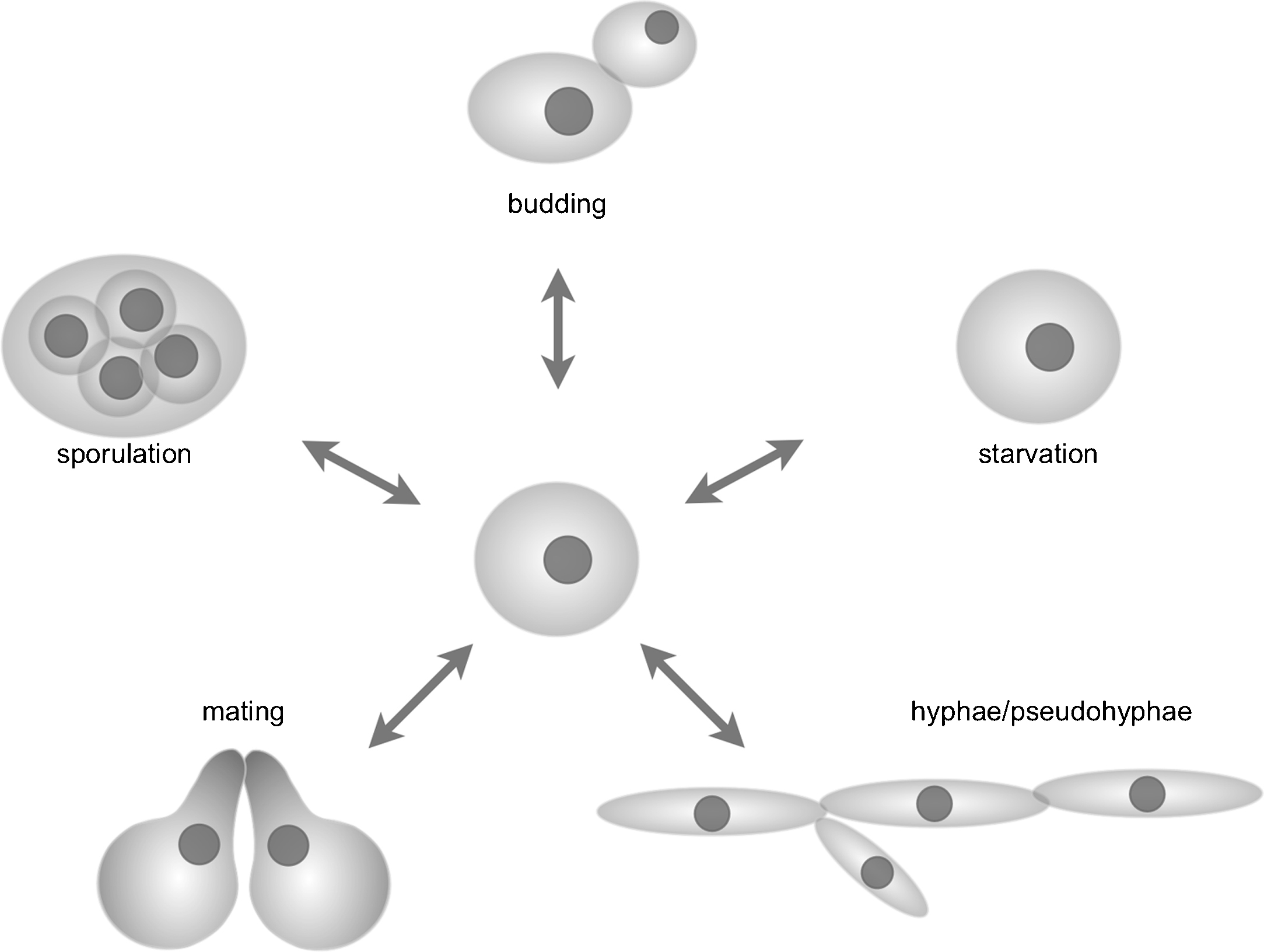
Developmental decisions in yeasts, exemplified by the well-known species S. cerevisiae. Decisions are either gradual, such as hyphae formation and starvation, whereas decisions that require a nuclear event (mitosis, mating and sporulation) involve a point-of-no-return beyond which the whole program has to be executed in order to return to a viable cellular state. Hence, correct developmental decisions are of crucial importance and therefore governed by sophisticated sensory mechanisms that integrate all the factors relevant for the decision.
Ploidy – The life cycles of yeast species typically involve haploid and diploid stages, separated by mating and meiosis/sporulation events, respectively (Fig. 3). Each species seems to have a preferred ploidy state - haploid or diploid - as inferred from the predominant ploidy state of isolates from the natural habitats of the species and from behavioral observations made in the laboratory. Judged from these observation S. cerevisiae is considered to be a diploid species, whereas Candida glabrata is haploid. In fact, for C. glabrata no diploid forms have been observed so far, although strains of both mating types have been reported [15]. In turn, S. cerevisiae grows well as haploid, but will return to a diploid state if mating is not prevented, e.g. by inhibiting mating type switching in clonal colonies. For many other species no prevalent ploidy state has been reported, which may indicate the existence of both ploidy states in the wild (summarized in [16]).
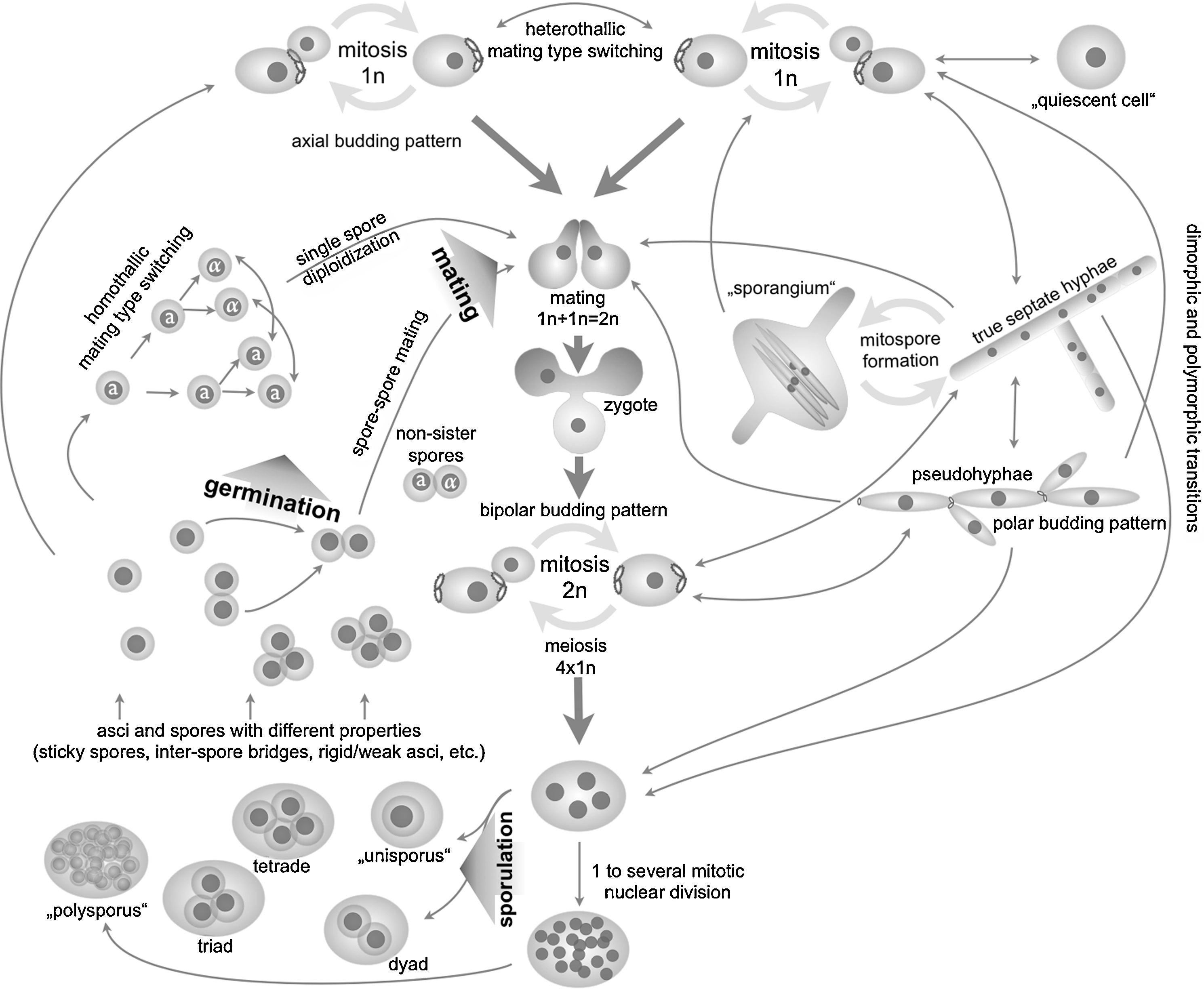
A generalized life cycle of the yeasts. Yeast life cycles are complex and contain cellular stages involving many different forms and shapes of cells. The scheme outlines the most prominent forms and the transitions between them. These are haploid growth phases, mating and zygote formation, diploid growth phases, sporulation and return to a haploid stage, germination of spores. Dimorphic transitions are also outlined: hyphal and pseudohyphal forms. For each of the transition the species have evolved individual sophistications that must have emerged as an evolutionary adaptation to a particular environment. Regulation of these transitions occurs globally but, after the decision has been made, involves several levels of regulation in order to specify the outcome to match as much as possible to specific parameters of the environment. Usually, these adaptations integrate internal signals, such as the available internal resources with external cues using a multitude of signal transduction and sensory pathways. The scheme integrates a broad range of observations made with different species. It thus does not report about the life cycle of an individual species, nor does it report on possible alternative solutions individual species may have evolved. Explanations for the different parts of the panel can be found throughout the manuscript.
Why are some species haploid and others diploid? From a survey of the evolution of the mating type loci and the HO endonuclease, which promotes active mating type switching (homothallism), the evolution towards an efficient way to change the mating type is apparent [17,18]. In clonal populations generated by one single cell or spore, it is clear that active mating type switching is a premise for an efficient return to a diploid life. The ability for active mating type switching has been invented more then once, and different mechanisms evolved, as shown by the comparision of the homothallic species S. cerevisiae and the fission ascomycetous yeast S. pombe [19,20]. Diploidy as a driving force for mating type switching is not evident, since S. pombe formally lives a haploid life. However, S. pombe cells spend most of the growth period in the G2 phase of the cell cycle and could hence be considered as pseudo-diploid. Mating, which is only stimulated by nutritionally sparse conditions, can consequently be seen as a necessary predecessor process for the immediately following entry into meiosis and spore formation.
What is the advantage of a diploid life style? Theoretical models discuss the advantages of diploidy in the context of deleterious mutations and their elimination via a sexual cycle [21]. Experimental evolution furthermore provided indication for an advantage of diploids during adaptive evolution [22].
5.1 Sporulation
Spore morphogenesis is tightly controlled, giving rise to highly differentiated structures with precisely specified shapes. S. cerevisiae spores are almost perfectly spherical. In contrast, the mitospores of A. gossypii are extremely elongated with two pointy ends. Other yeasts have reniform spores. Currently, not much is known about the molecular mechanisms underlying the generation of these shapes. Another parameter that is subject to control is the number of spores formed per cell per meiosis. S. cerevisiae and closely related species such as S. paradoxus are able to discard individual haploid genomes at the end of meiosis, because they can specify a subset of the 4 postmeiotic genomes for inclusion into spores. This is a sophisticated adaptive response to limited nutrients in the environment when the cell cannot afford 4 costly spore walls. This enables a perfect optimization of spore production, on the level of the individuals, but also on level of populations [23]. In the absence of this regulation spore formation has to be initiated at a time point when the cell is ‘sure’ about the availability of sufficient nutrients for 4 spores. Alternatively, constitutive abortion of 1–3 spores can occur, such as it appears to be the case in S. unisporus or Zygosaccharomyces bisporus. It has been shown for S. cerevisiae and other species that spore morphogenesis further considers the possibility of mating between a particular pair of spores, the so-called non-sister spores, which contain each a haploid genome derived from different homologous chromosomes (and not from sister chromosomes). In order to occur with high frequency, this requires that the mating type locus is linked to the centromere, in which case such pairs of spores are always of opposite mating type [16,24]. In S. cerevisiae and in Saccharomycodes ludwigii interspores bridges that connect pairs of spores have been observed [25,26]. In S. ludwigii these bridges form between non-sister spores and ensure direct mating of the associated germinating spores [27]. Whether this is also the case for S. cerevisiae remains open. Mating type linkage to the centromere and even some aspects of chromosome behavior in meiosis, such as the distribution and frequency of crossovers, are speculated to be influenced by high inbreeding rates between spores from related meioses, driven by the possibility to preserve genomic diversity and to complement efficiently lethal or deleterious mutations [9,28,29].
5.2 Mating
Mating denotes the formation of an asexual diploid cell from two sexual haploid cells of opposite mating type (MATa and MATα). This involves the fusion of two cellular cytoplasms, and the process is concluded with the fusion of the two nuclei (karyogamy). A mutual communication is required in order to execute mating. The nuclei committed to fuse need to be in the same stage of the cell cycle. To account for this, signaling of the mating response is limited to the G1 phase of the cell cycle by the cyclin dependent kinase (CDK) [30]. CDK inhibits the signal transduction machinery through which a cell senses the presence of a cell of opposite mating type. The subsequent mating response then leads to a halt in the cell cycle and to the execution of the steps leading to cell fusion.
S. cerevisiae haploid cells mate spontaneously, and it is not exactly known to what extent stress or starvation conditions will interfere with the mating process. Yeast mating involves the secretion of a soluble pheromone, the α-factor, from cells of the mating type MATα. Secretion of the α-factor occurs via the secretory pathway. Cells with the mating type MATa on the other hand use as a mating signal a fatty acid modified and therefore highly lipophylic peptide, the a-factor, which is cytoplasmically produced and excreted via an ABC-transporter. It can be hypothesized that the combination of a soluble pheromone (in Matα cells) with a lipophylic pheromone (in MATa cells) is a solution to many different mating scenarios. For example, S. cerevisiae has been shown to mate efficiently in liquid cultures as well as on solid support medium. Both conditions are diametrically opposed to each other, as they will generate entirely different effective pheromone levels. The a-factor will probably mostly reside in the plasma membrane of the excreting cell and hence serve as a ‘contact signal’, whereas the α-factor is secreted and thus able to signal over distances. Agglutination by surface lectins then leads to the formation of cell clumps. Yeast cells are able to polarize according to the direction from where the pheromone is emitted, and cell morphology adapts to the expected distance of the mating partner [31]. In dense cell populations such as cell clumps, high concentrations of pheromone are potentially reached due to the presence of many cells that actively produce the pheromones. A secreted protease specific for the α-factor resides in the cell wall and is thought to lower pheromone concentrations locally and thereby prevent saturation of the pheromone receptor, since this could potentially hinder the cell to read out the expansion of the pheromone gradient and to sense the position of the next mating partner [32] (M. Piel and A.W. Murray, personal communication).
5.3 Mating type switching
The mating type cassettes in the yeasts are thought to derive from functionalized genomic parasitic elements [33]. The evolution of mating type loci has been well studied [17,18]. It started with the establishment of the mating type specifiying genetic elements, the mating type cassettes. The acquisition of additional but silent copies of both mating type cassettes then permitted occasional mating type switching, thus providing the option for a return to a diploid life cycle in the absence of cells of opposite mating type. The further acquisition of a nuclease enabled active mating type switching, called homothallism, by regulated exchange of the active mating type cassette with the silent cassette of the other mating type. The level of sophistication that underlies this process has been well studied for S. cerevisiae [20].
5.4 Mating partner selection
Mating partner selection is a big issue for yeast and responsible for many of the morphological characteristics of each individual species. Mating partner selection in yeasts is not an active process but a passive mechanism that determines statistically whether a cell is mating with a related cell, e.g. a spore of the same ascus, mother-daughter mating or with another cell from the same cell clump. Nevertheless, in laboratory studies it has been found that yeast can evolve traits that specify the selection of preferred mating partners based on the mating fitness (or speed) [34]. To what extend this is relevant in the wild is not known. What matters really for yeasts are the levels of outbreeding and inbreeding during average mating events. This influences the effective population size and hence the potential to adapt to changes in the environment [35]. One factor influenced by cell morphology is the likelihood that related cells are next to each other when conditions favor mating. For example spores that are within a strong ascus tend to mate with each other upon germination, as seems to be the case for S. cerevisiae [23] or S. kluyveri, whose spores are very difficult to separate mechanically [36]. In contrast, other species have weak asci and the spores tend to disperse [37]. The shapes of spores may also influence dispersal of the spores via interactions with potential vectors (or hosts, in the case of pathogenic yeast species). Altogether, ascus and spore morphology determines the frequency of mating events between partners from the same meiosis (intra-tetrad mating), of mating between spores from related cells (adhering versus dispersing vegetative cells), and of mating between spores or the vegetative offsprings of unrelated cells (outbreeding/amphimixis enforced by a haploid life style due to absence of mating type switching).
5.5 Budding patterns
Budding patterns denote the position of appearance of a bud with respect to the position of the previous bud(s). Each budding event leaves behind a mark, the so-called bud scar that can be stained specifically using the fluorescent dye calcofluor white. Budding patterns have been characterized in S. cerevisiae [38] and shown to correlate with the ploidy state. Diploid patterns are either bipolar or polar, and the transition between polar and bipolar budding patterns is associated with the dimorphic transition. Haploids grow by axial budding, where each bud appears adjacent to the mark of the previous buds. This axial budding pattern is best explained in conjunction with the observed rules for active mating type switching that have been described earlier [39]. The outcome of active mating type switching in combination with axial budding in haploids enables a rapid and efficient diploidization. This synergism was proposed to stem from an evolutionary optimization for an efficient and quick return to a diploid life cycle [40]. Consistently, analysis of the molecular machinery underlying bud site selection indicates that random budding is the default behavior, whereas the axial pattern of haploids and the diploid budding patterns depend on specific genes and regulation imposed by the mating type [41].
6 Conclusion
Yeasts are a fantastic resource for discovery research and for developing and testing methods. The incredible richness of the species landscape, the ease with which they can be cultivated in the laboratory and the efficiency with which they can rapidly be converted into models that allow to use reverse genetics tricks for experimental testing of hypotheses make them an ideal playground for research on an evolutionary scale [42]. An example is RNAi in yeasts and it secondary loss in species like S. cerevisiae [43]. From the recent developments it now becomes more and more apparent that the future of basic research with yeast is no longer a di-species (S. cerevisiae and S. pombe) centric research. Instead, all yeast move to the center of focus and can now be chosen for their potential to investigate particular questions.
Disclosure of interest
The author declares that he has no conflicts of interest concerning this article.
Acknowledgements
Many thanks go to Andreas Kaufmann for the image of Ashbya gossypii and other valuable hints. Anton Khmelinskii is kindly acknowledged for valuable comments on the manuscript and Audrey Peltier for help with translation into French.


