1 Introduction
Annona muricata L. (Annonaceae), known as corossol, is a widespread small tree native in Central America, which has now a widespread pantropical distribution. The sample we studied was collected in the south of Senegal, where it is cultivated for its fruits. There, traditional medicine also uses this species, named ndélesor, in the treatment of many diseases, especially as antiseptic, healing substance and also against dermatosis and malaria fever [1]. In previous studies, we described the structure elucidation of cyclic peptides from Jatropha species, some of them having antimalarial activity [2,3]. Continuing our investigation of the cyclopeptides from plants, we here report on the isolation from the seeds of Annona muricata and the structural elucidation, based on tandem mass spectroscopy and 2D NMR, of the novel cyclic peptide, annomuricatin C (1).
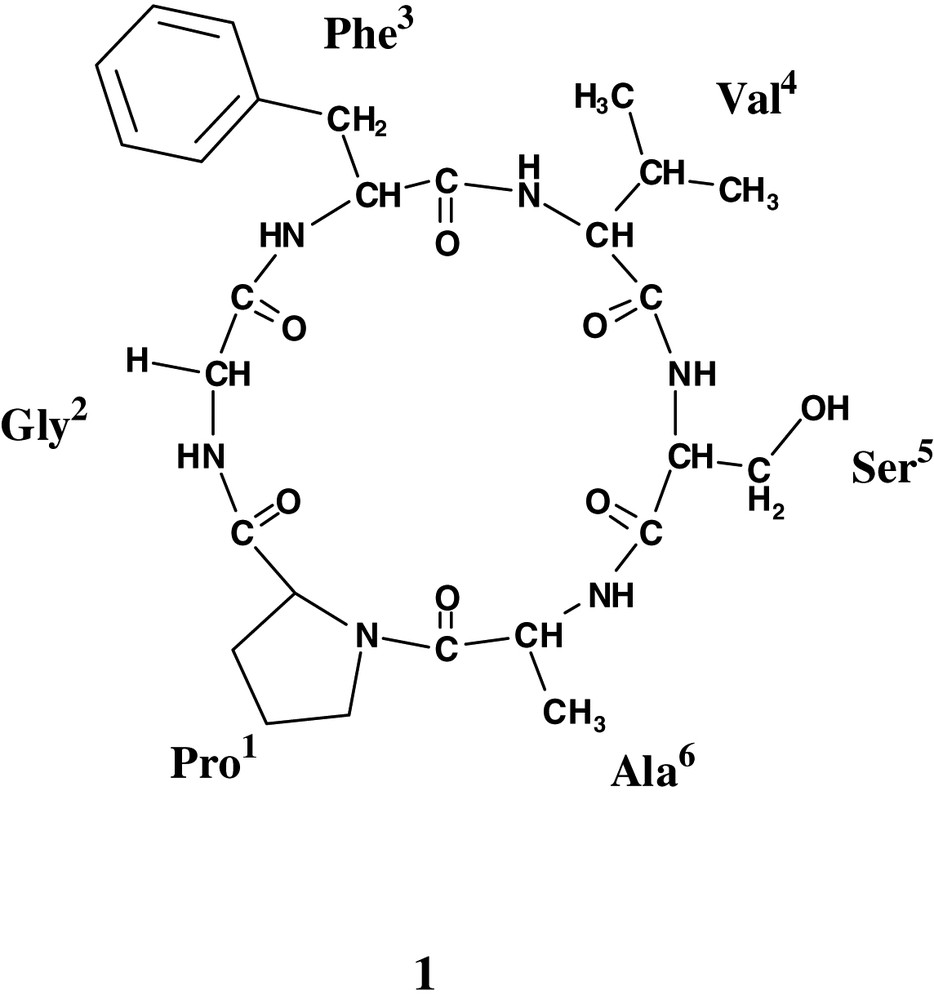
Most of the previous phytochemical studies on this species are related to acetogenins, as more than 50 compounds of this class were reported [4], and the two cyclopeptides annomuricatins A and B were also described [5,6]. Annomuricatin C was found to be cytotoxic against tumoural KB cells, with an IC50 1.0 μM.
2 Results and discussion
2.1 Isolation and characteristics
The seeds of Annona muricata were successively extracted with cyclohexane and MeOH and the MeOH extract after concentration was dissolved in EtOAc. The EtOAc soluble fraction was purified by exclusion chromatography, silica-gel column chromatography and C18 reversed-phase-HPLC to yield a new cyclopeptide termed annomuricatin C (1). Positive reaction with chlore/o-tolidine reagent suggested it was a peptide and the absence of coloration of its TLC spot with ninhydrin that it was cyclic. Analysis of the total acidic hydrolysate, after derivatization, indicated the presence of Ala (1), Gly (1), Phe (1), Pro (1), Ser (1) and Val (1). The amino acids were converted into the n-propyl esters of their N-trifluoroacetyl derivatives, analysed by gas chromatography on a chiral capillary column ; their retention times compared with those of standards indicated that all the chiral amino acids were l.
2.2 Mass spectral analysis
The molecular weight 558 for annomuricatin C (1) was deduced from the positive ESI-QTOF spectrum, which displayed the [M+Na]+ adduct ion at m/z 581 and the protonated molecular [M+H]+ ion at m/z 559. According to the amino acid composition, the molecular formula C27H38N6O7 was assigned to 1.
Cyclopeptides are not easily sequenced even by mass spectrometry. The reason is that multiple and indiscriminate ring-opening reactions occur during the CID of cyclic peptides, resulting in the superimposition of random fragment ions and making the interpretation difficult [7,8]. However we have shown that when a proline is present in the sequence, a specific fragmentation occurs at the peptidyl–prolyl (Xaa–Pro) amide bond, leading to a linear peptide C-ended by an acylium ion (bn), which undergoes further fragmentation, generating series of acylium ions from which the sequence could be deduced [2,9]. This specific fragmentation is explained by the more basic nature of the proline nitrogen relative to the other peptide-bond nitrogen atoms. In this way, the more basic site at proline level strongly directs the protonation, making the fragmentation less complex.
The protonated molecular ion [M+H]+ of 1 at m/z 559 was subjected to CID experiment (Fig. 1). The ring opening began at the Ala-Pro amide bond level and a series of adjacent acylium ions (bn) at m/z 488, 401, 302 and 155 was generated, from which the sequence could be deduced: amino acid residues were lost sequentially from the C-terminus to the N-terminus and for annomuricatin C (1) was observed the successive losses of Ala, Ser, Val, and Phe, yielding the N-terminal dipeptide Pro-Gly (Fig. 2). A second significant series of ions was observed at m/z 531, 460, 373, 274, 127 and 70, which were assigned to adjacent an ions related to the above bn ion series. At m/z 541 was observed an abundant ion originating from the loss of a molecule of water from the protonated molecular ion [M+H]+. It underwent fragmentation, yielding a third series of abundant ions here termed hn (Fig. 1B), corresponding to the successive loss of Val, Phe, Gly, Pro and Ala, giving a final ion at m/z 70, which corresponded to a dehydrated Ser residue. This latter series of adjacent fragments confirmed the above sequence and was explained by a favoured cleavage of the Val–Ser amide bond after Ser dehydration; this cleavage could be due to the formation of a cyclic amino acid residue from Ser.

CID fragmentation of the protonated annomuricatin C (1) ion.
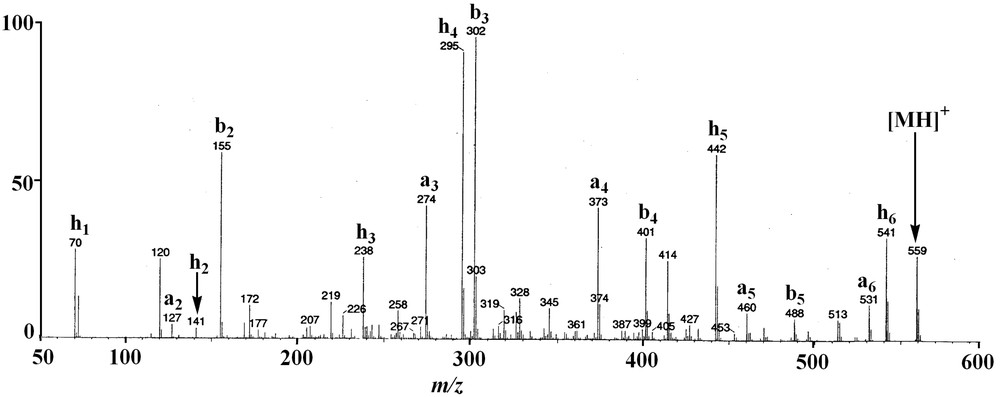
MS/MS fragmentation of the protonated annomuricatin C (1) ion: (A) fragmentation at the Ala–Pro amide bond level; (B) fragmentation at the Val–Ser amide bond level.
When analysing the fragmentation of the cationated adduct [M+Na]+ ion at m/z 581, it was observed that the energy collision needed for fragmentation should be higher (60 eV) and that the corresponding bn series was not observed. The main observed fragmentation yielded an abundant series of an ion fragments at m/z 553, 482, 395, 296 and 149, in agreement with the above sequence. The base peak at m/z 563 was due to the loss of a water molecule from the [M+Na]+ ion. These results suggested the sequence [H–Pro1–Gly2–Phe3–Val4– Ser5–Ala6]+ for the linearised peptide ion derived from annomuricatin C and thus the structure 1 for the natural cyclohexapeptide.
2.3 NMR study
The 1H-NMR spectrum of annomuricatin C (1) in DMSO-d6 solution showed a main stable conformational state (> 85%) for which the five amide protons were clearly depicted, as well as the presence of six carbonyl groups in the 13C-NMR spectrum, in agreement with a hexapeptide structure including a proline (Table 1). The peptide sequence determination was based on the HMBC experiment. This heteronuclear methodology was preferred to the homonuclear method described by Wüthtrich and based on dNN(i,i+1) and dαN(i,i+1) connectivities from the ROESY/NOESY spectra [10,11], because for small size cyclic peptides, conformational information can interfere with sequential ones. All the amino acid spin systems were identified using scalar spin–spin couplings determined from the 1H–1H COSY and TOCSY experiments [12]. The 13C-NMR assignments of the protonated carbons were obtained from the proton detected heteronuclear HSQC spectrum and combined with the HMBC experiment optimised for a long-range J-value of 7 Hz, for the non-protonated carbons. This experiment allowed the carbonyl groups to be assigned. By this way, the sequence determination was done from the connectivities between the carbonyl of residue i with the amide and/or α protons of residue i + 1 (Fig. 3). The 3JCH, CO (i) to NH (i+1) correlations depicted on the HMBC spectrum are shown in Fig. 4. As examples, the CO group of Gly2 at δ 168.0 was correlated to both αH and α'H of Gly2 and NH of Phe3, the CO of Ser5 at δ 168.8 to the αH and NH protons of both Ser5 and Ala6, and the CO of Pro1 at δ 171.8 to the α, α'-H and NH of Gly2 (Figs. 3 and 4).
13C and 1H NMR data for annomuricatin C (1) (DMSO d6, 318 K)
| Residue | δC | δH | m | J (Hz) | −Δδ/ΔT (10–3 ppm K–1) |
| Pro1 CO | 171.8 | — | |||
| α CH | 61.4 | 4.12 | dd | 5.6, 5.6 | |
| β CH2 | 28.7 | 2.09 | m | ||
| — | 1.83 | m | |||
| γ CH2 | 25.2 | 2.05 | m | ||
| — | 1.84 | m | |||
| δ CH2 | 47.0 | 3.77 | m | ||
| — | 3.49 | m | |||
| Gly2 CO | 168.0 | — | |||
| NH | — | 8.87 | dd | 8.6, 4.1 | 5.3 |
| α CH2 | 42.3 | 3.83 | dd | 17.1, 8.6 | |
| — | 3.31 | dd | 17.1, 4.1 | ||
| Phe3 CO | 169.7 | — | |||
| NH | — | 8.23 | d | 9.5 | 0.9 |
| α CH | 53.7 | 4.61 | m | ||
| β CH2 | 38.7 | 3.26 | m | ||
| — | 2.62 | dd | 12.7, 11.0 | ||
| 1′ | 137.5 | — | |||
| 2′, 6′ | 129.7 | 7.23 | m | ||
| 3′, 5′ | 127.9 | 7.21 | m | ||
| 4′ | 128.8 | 7.22 | m | ||
| Val4 CO | 170.8 | — | |||
| NH | — | 8.03 | d | 8.5 | 3.5 |
| α CH | 62.2 | 3.91 | dd | 9.8, 8.5 | |
| β CH | 29.4 | 2.07 | m | ||
| γ CH3 | 19.5 | 0.86 | d | 6.5 | |
| γ′ CH3 | 19.3 | 0.88 | d | 6.6 | |
| Ser5 CO | 168.8 | — | |||
| NH | — | 8.12 | d | 9.1 | 3.5 |
| α CH | 54.8 | 4.28 | m | ||
| β CH2 | 61.0 | 3.74 | m | ||
| — | 3.54 | m | |||
| Ala6 CO | 170.5 | — | |||
| NH | — | 7.42 | d | 7.3 | –0.3 |
| α CH | 46.3 | 4.59 | dd | 7.3, 6.9 | |
| β CH3 | 17.5 | 1.23 | d | 6.9 |

HMBC spectrum of annomuricatin C (1) CO regio-correlations.

(A) HMBC and (B) NOEs correlations for annomuricatin C (1).
The ROESY spectrum clearly showed only two dNN(i, i+1) interactions between Gly2 and Phe3, and between Ser5 and Ala6. A stretch of dαN(i, i+1) sequential connectivities was depicted from Pro1 to Ala6 (Fig. 4). The α proton of Ala6 gave a strong NOE correlation with the δ proton (3.77) of Pro1 and a weaker with the geminal δ' proton (3.49), closing the cyclohexapeptide ring and indicating that the Ala6-Pro1 amide bond was trans. Chemical shifts of the β and γ carbons of Pro1 at 28.7 and 25.2 ppm, respectively, gave further evidence for the presence of a trans-Pro amide bond [13]. The prochiral assignment of the β, γ and δ protons of Pro resulted of the observed NOEs in the ROESY spectrum. In addition to the sequential NOEs, some other interactions with conformational significance were depicted: the NH proton of Ser5 was strongly correlated to the β-proton of Val4, and the β-methyl of Ala6 to the δ'-proton of Pro. The whole data agreed with the cyclic structure 1 for annomuricatin A, the sequence of which was thus determined as cyclo(Pro1-Gly2-Phe3-Val4-Ser5-Ala6-).
Cyclic hexapeptide backbone rings are constrained with two β-turns generally stabilised by two transannular hydrogen bonds involving the two residues joining the β-turns and forming a short antiparallel β-sheet arrangement. Analysis of the NMR data of annomuricatin C suggested a constrained structure, leading to a main conformation.
All the 3JNH–Hα coupling constants had high values, between 7.3 and 9.5 Hz, in agreement with β-turn and β-sheet structures. In a β-turn consisting of four residues numbered i to i+3, a trans-Pro1 can only be located at the i+1 position, and as a result the following residue (Gly2) at the i+2 position. This is corroborated by the NOEs observed between NH–Phe3 (i+3 position) and both NH- and αH-Gly2, and between NH–Gly2 and αH–Pro1. This first β-turn with Pro1 and Gly2 at the corners has thus the characteristics of a type II β-turn (Fig. 5). The second β-turn has Val4 and Ser5 at the corners and can be defined as a type-I β-turn. This is deduced from the strong dNi, Ni+1 and dαi, Ni+1 interactions observed between Ser5 and Ala6 and the NOEs observed between NH-Ser5 and α-H and β-H of Val4. Confirmation of the conformation shown in Fig. 5, arose from the NOESY correlations between NH–Val4 and α-H and β-H of Phe3, and between NH–Ala6 and β-H of Phe3.
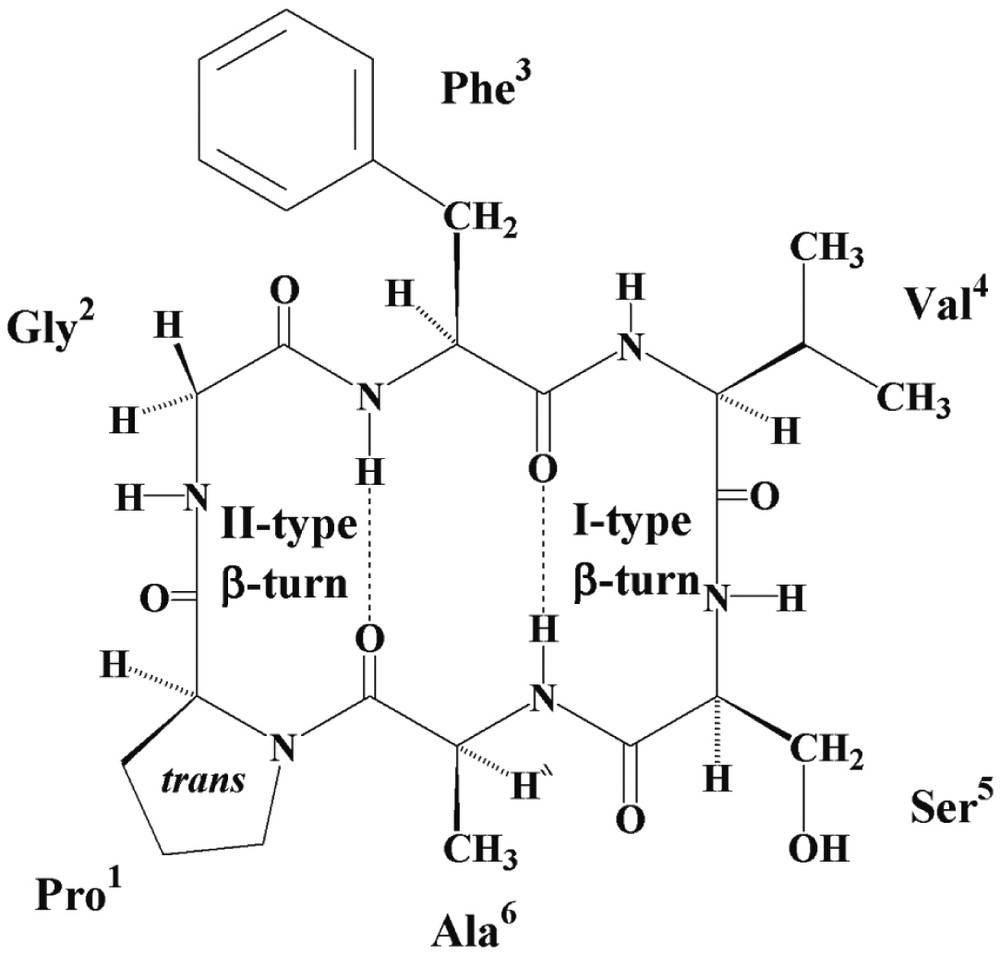
Solution conformation suggested for annomuricatin C (1).
The temperature coefficients Δδ/ΔT were measured between 298 and 328 K in DMSO-d6 (Table 1). The observed variations were linear and indicated that two amide groups (Phe3 and Ala6) with temperature coefficients between –0,9 and +0,5 × 10–3 ppm K–1 were involved in intramolecular hydrogen bonding, whereas the three other, especially that of Gly2 (Δδ/ΔT: 10–3 ppm K–1) were solvent exposed, in agreement with two hydrogen bonds between CO–Ala6 and NH–Phe3 and between CO–Phe3 and NH–Ala6. The conformation of annomuricatin C, as typically observed for the cyclohexapeptide rings, appeared to be formed by a short antiparallel β sheet, stabilised by two hydrogen bonding and two β-turns, a type-II turn depicted for Pro1-Gly2 and a type-I turn for Val4–Ser5 (Fig. 5).
It is remarkable that the peptides isolated from the Annonaceae family, hexa- to nonapeptides, enclose at least one proline residue. As usual, this residue induces, in such small-size cyclic peptides, conformational constraints. In addition, its presence is useful, as it favours the formation of one linearised peptide, the mass fragmentation of which allows the sequence to be unambiguously determined.
3 Experimental section
Optical rotation was measured with a Perkin-Elmer model 341 Polarimeter and the [α]D22 values are given in deg cm2 g–1. The melting point was determined on a Büchi melting point B-545 apparatus. 1H and 13C NMR spectra were recorded with either (1D 13C) a Bruker AC 300 spectrometer, equipped with an Aspect 3000 computer using DISNMR software or (2D spectra) a Bruker Advance 400 spectrometer operating at 400.13 MHz (2D spectra). The coupling constant used to establish the necessary delay for the selection of the proton coupled to the carbon in the HSQC spectrum was 135 Hz, corresponding to a delay of 3.7 ms; the delay for the HMBC spectra was 70 ms, corresponding to a long-range coupling constant of 7 Hz. The phase-sensitive ROESY experiments were obtained with mixing times of 150 ms. Mass spectra were recorded on a MALDI-TOF or an API Q-STAR PULSAR i of Applied Biosystems. For the CID spectra, the collision energy was 40 to 60 eV and the collision gas was nitrogen.
3.1 Plant material
Seeds of Annona muricata L. (Annonaceae) were collected near Dakar (Senegal) in January 2000. Samples were immediately washed with distilled water and dried at room temperature. A voucher has been deposed at the National Museum of Natural History (Paris).
3.2 Extraction and isolation
The dried and powdered seeds of A. muricata (1.3 kg) were macerated three times with cyclohexane (3 l) and the combined extracts yielded an oil (344 g) that was discarded. The seeds were then extracted three times with MeOH (3 l) at room temperature to give after concentration under reduced pressure the MeOH extract (89.7 g), which was partitioned between EtOAc and water. The organic phase was concentrated to dryness and the residue (15.8 g) was dissolved in MeOH and chromatographed on Sephadex LH-20 column with MeOH. The peptide fraction (15.8 g) was then repeatedly subjected to silica gel column chromatography (Kieselgel 60 H Merck) and eluted with CH2Cl2 containing increasing amount of MeOH from 5% to 20%.
The peptide purification was monitored by TLC (silica gel 60 F254 Merck) with CH2Cl2/MeOH 9:1 as eluent system and the peptides were detected with the Cl2/o-tolidine reagent, exhibiting a characteristic blue spot with Rf 0.38. The corresponding peptide mixture was finally purified by isochratic reversed phase HPLC (Kromasil C18, 250 × 7.8 mm, 5 μm, AIT France; flow rate 2 ml min–1, detection 220 nm) using MeOH/H2O: 65/35 with 1% TFA to yield annomuricatin C (1, tR 7.2 min, 23.1 mg).
3.3 Absolute configuration of amino acids
Solution of 1 containing 1 mg of peptide, in 6 N HCl (1 ml), was heated at 110 °C for 24 h in sealed tubes. After cooling, each solution was concentrated to dryness. The hydrolysate was dissolved in anhydrous solution of 3 N HCl in 2-propanol and heated at 110° C for 30 min. The reagents were evaporated under reduce pressure and the residue dissolved in CH2Cl2 (0.5 ml) and 0.5 ml trifluoracetic anhydride was added. The mixture was kept in a screw-capped at 110 °C for 20 min, the reagents were evaporated and the mixture analysed on a Chirasil-l-Val (N-propionyl-l-valine-tert-butylamide polysiloxane) quartz capillary column with helium (1.1 bar) as carrier gas and temperature program of 50–130 °C at 3 °C min–1 and 130–190 °C at 10 °C min–1, with a Hewlett Packard series 5890 apparatus. Comparison of Rt values with those of standards amino acids was used: l-Ala (11.6), l-Val (13.9), Gly (14.6), l-Thr (15.2), l-Pro (18.2), l-Ser (18.8), l-Leu 19.2) and l-Tyr (31.9).
3.4 Annomuricatin C (1)
Colourless solid, Mp 284–285 °C (MeOH), [α]D22–2.7° (c 0.1, MeOH) ;1H and 13C NMR (see Table 1); ESI-QTOF, m/z: 597.23 [M+K]+, 581.26 [M+Na]+, 559.28 [M+H]+; ESI-QTOF MS/MS on m/z 559 [M+H]+ (ce 40 eV) m/z (%): 559 (28), 541 (34), 531 (12), 513 (9), 488 (7), 470 (4), 460 (9), 442 (61), 427 (4), 414 (28), 401 (33), 374 (11), 373 (44), 345 (10), 328 (13), 326 (9), 319 (10), 302 (100), 295 (96), 274 (44), 258 (11), 238 (27), 226 (7), 219 (12), 172 (10), 155 (61), 141 (4), 127 (5), 120 (26), 72 (13), 70 (30). ESI-QTOF MS/MS on [M+Na]+ (ce 60 eV) m/z (%): 581 (96), 563 (100), 553 (93), 535 (73), 523 (49), 507 (20), 492 (42), 482 (75), 466 (51), 464 (49), 454 (26), 436 (36), 422 (17), 405 (4), 396 (25), 395 (95), 368 (21), 367 (93), 324 (22), 306 (7), 297 (20), 296 (91), 276 (13), 234 (11), 220 (6), 202 (6), 163 (4), 149 (2).
Acknowledgements
The French ‘Ministère de la Coopération’ (EGIDE) is gratefully acknowledged for a fellowship for one of us (AW), and the ‘Région Île-de-France’ for its generous contribution to the funding of the 400-MHz NMR and the ESI-TOF mass spectrometers. We wish to thank M. A. Blond and M. L. Dubost for NMR and mass spectra contributions, respectively.


