1 Introduction
The human population inhabiting the coastal areas expands continuously (Cravo et al., 2015; Dorsey et al., 2013) and creates environmental stress on adjacent ecosystems. In this context, coastal ecosystems are particularly vulnerable. It is well known that human activities can influence the proliferation and distribution of fecal bacteria including potential pathogens (Cravo et al., 2015; Dorsey et al., 2013). Beside fecal indicators, the abundance and richness of antibiotic (ATB)-resistant bacteria in ecosystems are of major concern in the current era of global resistance burden (Laxminarayan et al., 2013).
At the “omics” era, metagenomic methods approach the stock of genes encoding ATB resistance in ecosystems (Amos et al., 2014; Kristiansson et al., 2011; Marti et al., 2014; Port et al., 2014; Schlüter et al., 2008; Szczepanowski et al., 2008). These culture-independent tools have demonstrated the huge diversity of resistance genes and encoded mechanisms, which form the environmental resistome. However, resistance to ATBs is a bacterial behavior observed in vitro as a resistant phenotype, which is predictive, in medicine, of patient treatment failure. Clinically resistant phenotypes are defined by comparison of Minimal Inhibitory Concentrations (MIC) values (concentrations that inhibit the growth of a bacterial strain) with thresholds called clinical breakpoints edited by expert committees for each ATB and bacterial type. When MIC is over the clinical breakpoint value, the strain is categorized as resistant to the tested ATB. Similarly, for environmental bacteria, in vitro studies of ATB-resistant phenotypes are generally performed on isolated strains of well-known pathogenic species (Maravić et al., 2012; Okoh and Igbinosa, 2010; Ramírez Castillo et al., 2013). Studies on isolated environmental autochthonous non-pathogenic bacteria are scarcer (Schreiber and Kistemann, 2013; Varela et al., 2016; Vaz-Moreira et al., 2011), and antimicrobial resistance of the whole cultivable communities is even less studied (Varela et al., 2014; Weber et al., 2011). Moreover, indicators are needed to evaluate both the vulnerability of ecosystems facing antimicrobial constraints and the potential risks for humans to be exposed to environmental ATB-resistant bacteria. In this context, aquatic environments are particularly interesting because they are involved in the dispersal and mix of antibiotics (and more generally xenobiotics) and antibiotic/xenobiotic-resistant bacteria from diverse origins (Baquero et al., 2008; Gaze et al., 2013).
While antibiotic resistance is monitored in human or animal medicine by standardized methods, there is still a need for harmonization to monitor antibiotic resistance in environment (Brauner et al., 2016; Rizzo et al., 2013). This study intends to propose such a method. We introduce an original approach to evaluate the resistance to antibiotics of bacterial communities in water systems. We report this approach through a pilot study of Mediterranean hydrosystems in the Montpellier region, France, where we analyzed both freshwater and brackish waters subjected to different levels of anthropization.
2 Materials and methods
2.1 Study area, water sampling and processing
We selected three sites with contrasted environments (Fig. 1). Two sites are in the Montpellier urban area (about 400,000 inhabitants) located in the Mediterranean coastal basins of the Gulf of Lion, France. Montpellier city is located in a small watershed that flows towards the Lez River. This coastal 13-km-long river runs through the city where it is channelized, and emerges in lagoons connected to the Mediterranean Sea such as the Mauguio Lagoon. The Lez catchment covers 709 km2 and contains the densely urbanized area of Montpellier. The first sampling site is located in the Lez River downstream the city (freshwater) (43.570878, 3.897733). The second is situated in the Mauguio Lagoon (43.554166, 3.970255) (brackish water). These two sites are subjected to strong anthropogenic pressure from the Lez watershed and the city of Montpellier. We selected a third sampling site in the Salagou Lake (43.654424, 3.384781), a preserved recreational site, classified Natura 2000 (Conservation of natural habitats of wild fauna and flora) and hence little anthropized compared to the other two sites.

Study areas and sampling stations in the Montpellier region (South of France) (géoportail: https://www.geoportail.gouv.fr/).
In March 2010, we collected water samples (1 L) at these three sites, in sterile glass bottles, 30 cm under the water surface. The samples were transported in coolers to the laboratory and processed within 4 h after their collection. One milliliter of each water sample was filtered across a 0.45 micron-pore-size nitrocellulose sterile filter (47 mm in diameter, Sartorius, Germany) using a vacuum filter apparatus. Bacterial communities retained by the filter were then resuspended in sterile water.
2.2 Determination of community Inhibitory Concentrations (c-IC) of antibiotics
For every water sample, we spread 100 μL of non-diluted and diluted (10 and 100 fold) bacterial community suspensions on Trypticase Soy Agar plates (TSA, Difco laboratories, USA) supplemented or not with each ATB at each tested concentration: cefotaxime (CTX, from 128 to 0.0625 mg/L), ceftazidime (CAZ, from 128 to 0.0625 mg/L), and tetracycline (TET, from 32 mg/L to 0.0625 mg/L). For brackish water samples, c-ICs were additionally measured onto Marine Agar (MA media, salted by 19.45 g/L of NaCl; Difco laboratories, USA). The MA media were treated with the same ATB concentrations as before.
Three ATBs were tested. Tetracycline (TET) is one of the more prevalent and persisting ATB in various environmental compartments (Tello et al., 2012). Two third-generation cephalosporins, cefotaxime (CTX) and ceftazidime (CAZ), are markers for innate or acquired multidrug resistance in major human pathogens like Pseudomonas aeruginosa and Enterobacteriaceae. Despite they belong to the same family, these two ATBs present different antimicrobial activities profiles.
For each medium and each ATB, a set of plates was incubated during 48 h at 20 °C, another set being incubated during 48 h at 30 °C. The temperature of 20 °C represents the average annual temperature at the sampling sites (from 6 to 23 °C). Temperatures below 20 °C were not used because they would slow bacteria growth and thus require longer incubation times, with the risk of degradation of the antibiotics. Incubation at 30 °C was chosen because it promotes the growth of potential human-associated bacteria. The enumeration of bacterial colonies (Colony Forming Units [CFU]) grown on media with and without ATBs allows us to define the c-IC50 and c-IC90 criteria, which correspond to the ATB concentration that succeeds in inhibiting 50% and 90% of the growth of the total cultivable community (CFU counted on media without ATB), respectively. These c-IC criteria can then be compared to the clinical breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2017) and the Antibiogram Committee of the French Society for Microbiology (CA-SFM, 2013). Clinical breakpoints used in this study are 2 mg/L for CTX, 8 mg/L for CAZ (EUCAST, 2017) and 8 mg/L for TET (CA-SFM, 2013).
2.3 DNA analysis to identify the taxonomic content of the bacterial communities
We used an enzymatic method (MasterPure Gram positive DNA purification kit, Épicentre, France) to extract the DNA from the total bacterial community growing onto TSA and MA media. DNA extracts were used as templates for amplification by polymerase chain reaction (PCR) of the V3 variable region of the 16S rRNA gene using the primers HDA1GC-HDA2 (Ogier et al., 2002). Temporal Temperature Gel Electrophoresis (TTGE) migration allows the separation of every 16S rRNA gene amplification product corresponding to every Operational Taxonomic Unit (OTU) forming the community. TTGE experiments were performed as described in prior works (Michon et al., 2010).
For OTU affiliation, TTGE bands were compared to diversity ladders or were sequenced. We constructed three diversity ladders, each corresponding to the Salagou Lake, Lez River, and Mauguio Lagoon community, as previously described (Roudière et al., 2009). Loaded on TTGE migration gel, comparison of TTGE bands to diversity ladders allowed the direct assignation of a band to a bacterial taxon. Taxon affiliation was also performed by sequencing DNA from selected TTGE bands excised from the migration gel (Michon et al., 2010) on an ABI 3730xl sequencer (Cogenics, UK). We compared sequences to GenBank and RDPII databases (Cole et al., 2014) for affiliation to an OTU considering the phylotaxonomic frame surrounding the OTU in RDPII. The crude taxonomic diversity index (DI) corresponded to the number of different bands per TTGE profile.
2.4 Statistical analyses
To compare the bacterial abundances and the levels of resistance to antibiotics of bacterial communities, we used the R software (available at http://www.R-project.org) and the R package FactoMineR (Husson et al., 2008). The normality of distributions and the equality of standard deviations were tested using Shapiro-Wilk and Fisher tests, respectively. The Student and Mann-Whitney tests were used to define statistical significant differences.
3 Results
3.1 Abundance of cultivable communities in coastal hydrosystem versus ATB selection
To explore the antimicrobial resistance at the bacterial community level, the freshwater bacterial communities were cultivated in 74 different conditions (TSA medium without or with various ATB concentrations using two incubation temperatures) and the brackish communities in 148 conditions (74 conditions as for freshwater samples but for both TSA and MA media). On ATB-free TSA medium, the abundance of cultivable bacteria was found similar for water samples from the Lez River (1.35·103 ± 2.97·102 CFU/mL) and the Mauguio Lagoon (2.15·103 ± 9.21·102 CFU/mL), but lower in the Salagou Lake's water (2.75·102 ± 1.10·102 CFU/mL respectively). The abundance of halotolerant/halophilic lagoon bacterial community (HHLC) growing on MA medium (3.37·103 ± 2.09·103 CFU/mL) was not significantly different from that observed on the TSA medium, corresponding to the total cultivable community called “non-halophilic lagoon community” (non-HLC) (p > 0.05). No significant differences in bacterial counts were observed between incubation runs at 20 °C and 30 °C (p > 0.05) (inserts in Fig. 2).
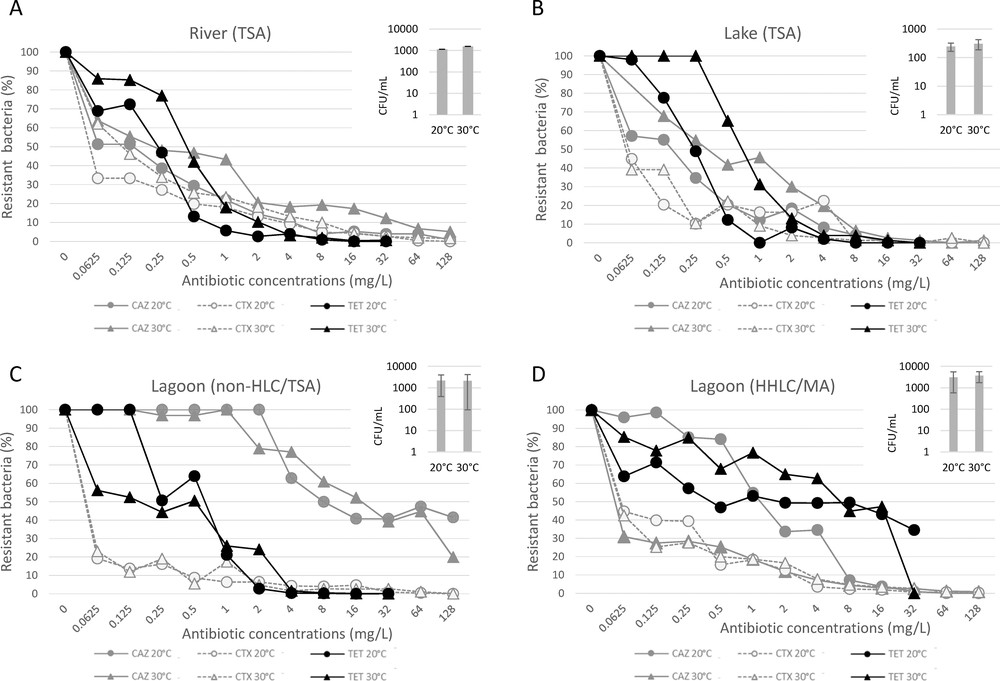
Percent of resistant cultivable bacteria in water samples from the Lez River (A), Salagou Lake (B), and Mauguio Lagoon (C, D) according to culture conditions (temperature and salinity) and antibiotic concentrations. In insets (top right of graphs), abundance of bacterial community in each sample, in CFU/mL, according to temperature of incubation. TSA: Trypticase soy agar; MA: marine agar; non-HLC: non-halophilic lagoon community; HHLC: halotolerant/halophilic lagoon community; CAZ: ceftazidime; CTX: cefotaxime; TET: tetracycline.
We then counted the bacterial communities grown at 20 °C and 30 °C onto media containing increasing concentrations of CTX, CAZ or TET. For both the coastal river waters and the continental lake freshwaters (Fig. 2A and B), the curves of abundance show roughly similar patterns whatever the temperature or ATB concentration. As expected, the main tendency is a decrease of abundance as the ATB concentrations increase.
By contrast, the resistance to CAZ of the brackish lagoon community is different from its resistance to TET (Fig. 2C and D). The amounts of bacteria resistant to TET and CAZ markedly differ depending on the salinity of the culture medium (TSA or MA) (p = 0.0119 and p < 0.0001, respectively) (Fig. 2C and D). The HHLC community is more resistant to TET than the non-HLC community. The proportion of the non-HLC resistant bacteria to TET, CAZ or CTX is not influenced by the temperature of growth (p = 0.6501, p = 0.9085, and p = 0.9548, respectively), but the HHLC community growing at 20 °C appears more resistant to CAZ than the equivalent community growing at 30 °C (Fig. 2D) (p = 0.00357). Conversely, the HHLC community growing at 30 °C is more resistant to TET than is the equivalent community growing at 20 °C (p = 0.0315). The curves of abundance of CTX-resistant bacteria in the Mauguio Lagoon show similar patterns whatever the salinity (p = 0.3012) and temperature (p = 0.9492).
Finally, overall, the Mauguio Lagoon bacteria communities show a higher resistance to ATB than the freshwater communities, with significant differences between the HHLC and the non-HLC sub-communities.
3.2 Assessing the resistance level of the cultivable bacterial communities from the c-IC50 and c-IC90 criteria
We then determined the c-IC50 and C-IC90 criteria. We observed that, for every type of water and in most conditions, low concentrations of ATBs (0.06 to 0.250 mg/L) inhibit the growth of 50% of the bacterial community (Fig. 3). This inhibition is particularly clear with CTX. Low ATB concentrations thus have a strong impact on the overall bacterial community structure. Concentrations that inhibit 90% of the community growth are markedly higher, generally ranging between 1 and 64 mg/L (Fig. 3).
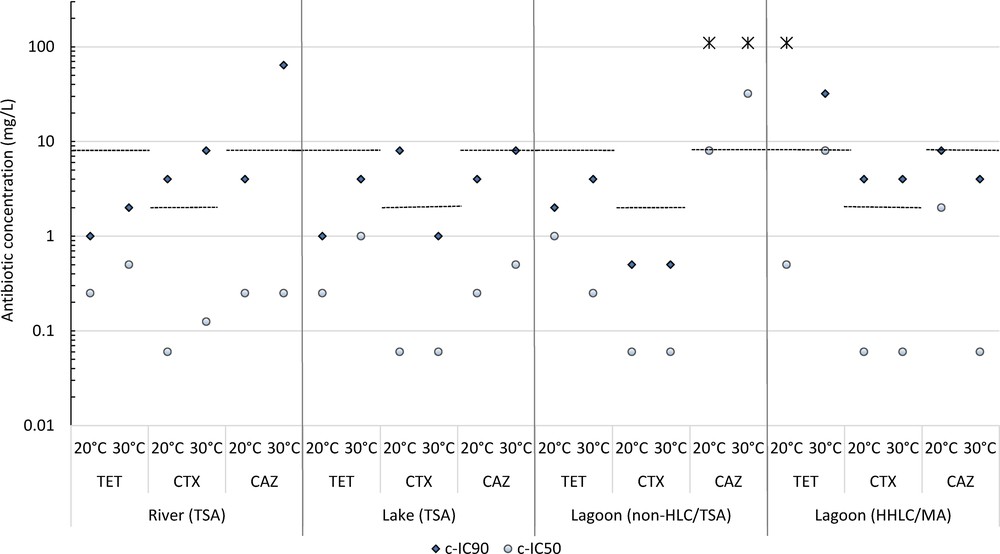
Concentration of antibiotics inhibiting the growth of 50% (c-IC50) and 90% (c-IC90) of the bacterial community. The stars indicate c-IC90 over the highest concentration tested. The dotted lines indicate clinical breakpoints for resistance defined by EUCAST and CA-SFM guidance; in clinical context, if MIC (minimal inhibitory concentration) is over 2 mg/L for CTX and 8 mg/L for CAZ and TET, it means that studied strain is resistant to the ATB tested.
For both freshwater samples, c-ICs are of the same order of magnitude and vary similarly with temperature conditions (Fig. 3). Bacteria communities from the brackish lagoon (HHLC and non-HLC growing at 20 °C or at 30 °C) show more contrasted c-IC profiles, varying with salinity and temperature conditions. As observed for the abundance results, the resistance level of the brackish lagoon communities assessed from the c-IC criteria is generally higher than that of the freshwater communities. For instance, the c-IC90 of CAZ exceeds 128 mg/L for non-HLC, while the c-IC90 of TET is also greater than the highest tested concentration, i.e. 32 mg/L at 20 °C for HHLC (Fig. 3).
The c-ICs criteria allow comparing the resistance of bacterial communities in the environment to those in clinical tests. In half of the conditions analyzed here, the c-IC90 is similar or greater than the clinical resistance breakpoints (Fig. 3). For HHLC, concentrations similar or greater than clinical breakpoints succeed in inhibiting no more than 90% of the bacterial community, whatever the ATB. The same observation is made for freshwater communities for cephalosporins (CTX and CAZ), but not for tetracycline (TET).
These results show that a significant part of the bacterial communities in natural surface waters can grow even though the concentrations in ATBs are higher than the threshold values used to define the bacterial resistance in medicine. This shows that natural waters are reservoirs of bacteria that have developed a resistance level to ATB in such a range that it could impair antimicrobial therapy in human or animals. Overall, the Mauguio Lagoon bacterial community shows a different resistance behavior than the community of its tributary, the Lez River. The level of resistance in the Lez River is similar to that in the Salagou Lake, despite the two zones are hydrologically unrelated and sustain different anthropization. We infer that salinity is a major factor that influences the overall resistance of aquatic bacterial communities to antimicrobial agents.
3.3 Taxonomic diversity in antibiotic-resistant cultivable communities
We established the repertory of OTUs among the communities growing in presence of ATBs by 16S rDNA PCR-TTGE fingerprinting. We chose to focus on ATB concentrations that induced changes in the shape of the curves in Fig. 2. The affiliation of a band to a taxon was done for 645 TTGE bands (79.2%) out of the 814 observed in our experiments, either by sequencing or by comparison with diversity ladders. The taxonomic diversity of ATB-resistant bacteria corresponds to a total of 33 genera (including five unidentified genera in Enterobacteriaceae and Vibrionaceae) and four phyla: Proteobacteria (56%), Firmicutes (17%), Bacteroidetes (17%) and Actinobacteria (10%) (see Supplementary data, Table S1). Fig. 4 compares the three communities in terms of their taxonomic repertory and resistance. Despite their hydrological continuity, the Lez River and the Mauguio Lagoon have only four genera in common, which are also found in the Salagou Lake: Bacillus, Pseudomonas, Shewanella, and Vibrio (Fig. 4A). Only one other genus, Rheinheimera, is common to freshwaters (lake) and brackish waters. Most genera (22/33) are specific to a given type of water: 11 in the lagoon, six in the river, five in the lake, and six in both the river and the lake freshwaters (Fig. 4A).
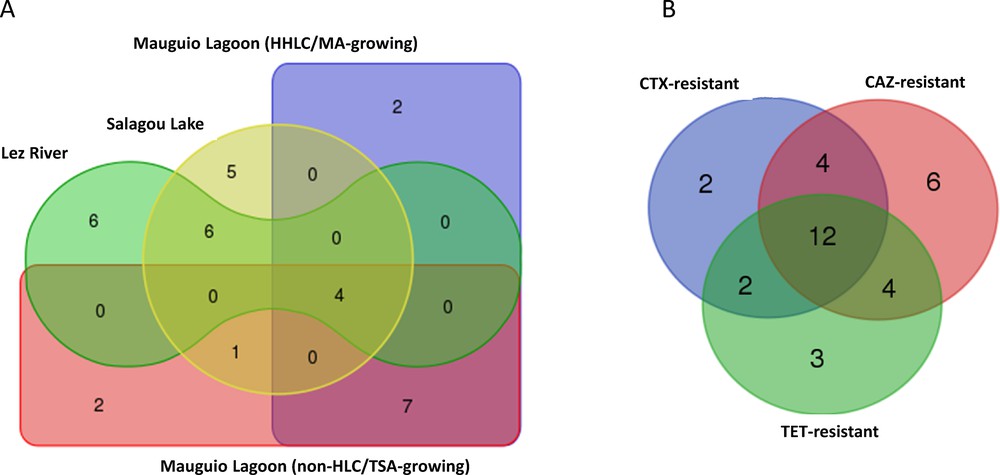
Venn diagrams showing the distribution of genera according (A) to the type of water and to the culture medium used; (B) to the resistance to ATBs; CAZ: ceftazidime; CTX: cefotaxime; TET: tetracycline.
We examined the distribution of genera versus their antimicrobial resistance (Fig. 4B). The genera that resist to CTX only are few (n = 2) while, frequently, CTX-resistant genera are also resistant to CAZ (n = 16) (Fig. 4B). Cross-resistance between the cephalosporins (CTX and CAZ) and TET is observed for 12 genera, compared to four genera displaying resistance to both TET and CAZ and two genera displaying resistance to both TET and CTX (Fig. 4B). Genera displaying a cross-resistance to the 3 ATBs (Table S1) are Pseudomonas, Rheinheimera, Vibrio, Flavobacterium, Cellulophaga, Marinibacillus, and Microbacterium from the lagoon samples, and Acinetobacter, Pseudomonas, Vibrio, Chryseobacterium, Elizabethkingia, Bacillus, and Exiguobacterium from the river samples. In all cases, the Alpha- and Beta-proteobacteria (Brevundimonas, Agrobacterium, Chromobacterium, Comamonas) are detected only when a cephalosporin is added to the culture medium. This suggests that these bacteria are minor in the communities, but are easily selected by antimicrobial pressure (i.e. under inhibition of growth of the originally dominant bacteria). CTX-resistance is observed for four genera in four families in the lake cultivable community, while this resistance is more widely distributed in coastal hydrosystems. Nine genera from eight families for Lez River and 14 genera from 12 families for Mauguio Lagoon present this resistance (Table S1 and Fig. 4B). Most genera with isolated TET-resistance belong to gamma-proteobacteria.
3.4 Dynamics of taxonomic richness and diversity with antibiotic concentration
The abundance of every bacterial family varies with the nature and the concentration of ATBs (Supplementary data, Fig. S1). These variations reveal the qualitative dynamics of the communities under antimicrobial pressure. In brackish waters, families are diverse and stable whatever the concentration of ATBs (Fig. S1A). In all waters, depending on the ATB concentration, some families become dominant up to be the sole representative of the cultivable bacterial community. It is mainly the case for Bacillaceae in freshwaters in the presence of the highest concentrations of cephalosporins. Several other families dominate the community for a range of ATB concentrations, and then decrease under higher concentrations. This tendency is particularly observed for Aeromonadaceae in the Lez River samples under TET pressure, and for Moraxellaceae in the Salagou Lake community under CTX selective pressure (Fig. S1B and C, respectively). Finally, several minor families are detected only in presence of an ATB. It is noteworthy that Vibrionaceae are selected in the Lez River water at high ATB concentrations only (Fig. S1B), whereas, in the Mauguio Lagoon, they are selected at most ATB concentrations (Fig. S1A).
The phylum Proteobacteria (green and brown in supplementary Fig. S1) displays the wider diversity of families in the three types of water. Some proteobacterial families are mainly specific for a type of water and for a type of ATB pressure. Moraxellaceae (mainly the genus Acinetobacter) dominates the lake community under CTX pressure, while Enterobacteriaceae and Aeromonadaceae are dominant under TET pressure (Fig. S1C). The two latter families also dominate the river water community under TET pressure (Fig. S1B). However, the river community is more diversified than the lake community, with a wider representation of members of the phylum Firmicutes (blue in Fig. S1B and C) beside Proteobacteria. Firmicutes, mainly the order Bacillales, are particularly selected under CAZ pressure in the river and lake waters (Fig. S1B and C). In brackish waters, the dominant family is Pseudomonadaceae whatever the ATB selective pressure, and this family shows a higher susceptibility to CAZ. The Mauguio Lagoon waters present the wider family diversity, with an overall persistence of taxon diversity as ATB concentrations increase. As a matter of fact, the persistent detection of Bacteroidetes is a particularity of the lagoon waters. It is also the case, but at a lesser extent, for members of the family Microbacteriaceae in the phylum Actinobacteria (Fig. S1A).
In conclusion, the structure of the bacterial communities in the freshwater samples is modified under ATB pressure, whereas the brackish-water community appears more stable and hence likely less vulnerable to antimicrobial agents. Moreover, bacterial families that are involved in human and animal infections, such as Enterobacteriaceae, Moraxellaceae (Acinetobacter), Aeromonadaceae, and Vibrionaceae, become dominant when exposed to ATBs, even at low concentrations.
4 Discussion
While there have been numerous studies on microbial communities in marine waters and sediments, freshwater and brackish coastal bacterial communities have been hardly studied so far (Wang et al., 2012), even though they are a major element of the ecosystems (Aliaume et al., 2007). In comparison to the available powerful metagenomic tools (Walsh et al., 2011; Xiong et al., 2015), culture-based studies are currently somehow disregarded to explore the bacterial communities, in part because they are unable to decipher the uncultivable microbial reservoir (Garcia-Armisen et al., 2013). Consequently, at present, culture-based approaches in water microbiology are mainly restricted to the count of a few microorganisms, such as E. coli and enterococci as indicators of fecal contamination (Rizzo et al., 2013). However, culture-based approaches have several advantages, as they allow the characterization of phenotypic susceptibility to antimicrobials, which is until now a major medical parameter predictive of treatment efficiency and clinical evolution (Köser et al., 2014). Moreover, even if the prevalence of resistance genes is assessed by culture-independent methods (Walsh et al., 2011; Xiong et al., 2015), a culture-based approach is needed to allow the characterization of the strains harboring these resistance genes. Furthermore, selective pressure could be exerted upon bacterial culture, thereby allowing rare populations to become dominant and these rare resistant populations are of major concern in human health (Huang et al., 2016).
Currently, culture-based studies of water bacterial communities are generally focused on fecal bacteria or on well-known human pathogens. However, every bacterium in an ecosystem can be involved in the spread of resistance genes, acting either as a reservoir or as a vector (Baquero et al., 2008). The main advantage of the approach developed in this study is to assess the whole cultivable bacterial community. However, in such cultivation-based studies, culture conditions are critical and might be a major bias. We observed that physical and chemical conditions (temperature and salinity) influenced the abundance of cultivable bacteria in presence of ATBs and thus the c-ICs. This indicates that preliminary studies involving a wide range of culture conditions are necessary on each type of water sample in order to cover most of the growth requirements of the bacteria present in the studied community. Culture conditions could also be chosen to reproduce as closely as possible the environmental conditions and/or to address specific questions, such as here, the resistance to antibiotics. In this context, development of culturomics approaches could be of great interest to enlarge the repertory of cultivable bacteria from diverse water samples (Diop et al., 2016). Of note, most of the ATB concentrations used in this study largely exceed those currently encountered in natural environments (Carvalho and Santos, 2016) because one aim of our study was to test hydrosystems bacterial communities as possible reservoirs for clinically relevant ATB-resistant bacteria.
We have focused here on three ATBs. Tetracycline is a major indicator of bacterial resistance in the environment because it is prevalent and persisting in those environments (Tello et al., 2012). The two other ATBs, CTX and CAZ, are third-generation cephalosporins (β-lactamins class), which are today used for curing life-threatening infections. We showed in vitro that concentrations of CTX from 0.0625 to 0.5 mg/L have a significant influence on both the abundance and the community structure of cultivable bacteria in surface waters. Moreover, pathogens such as Acinetobacter spp. (Moraxellaceae) can become selected in the presence of 2 mg/L of CTX (clinical breakpoint for resistance).
Resistance to CAZ has been investigated particularly because P. aeruginosa and Enterobacteriaeae resistant to CAZ are considered as emerging resistant pathogens. For these bacteria, resistance to CAZ defined multidrug resistance which is a major cause of treatment failure, mainly in the context of healthcare-associated infections (Oliver et al., 2015). Pseudomonas isolates resistant to CAZ were found in the three types of water that we studied.
The three tested ATBs revealed the in vitro selection of genera with potential impact on human infectious diseases. Combining c-IC determination with species identification can be a valuable improvement of the method to assess more precisely the infectious risk associated with the resistant genera selected in natural waters (Sala-Comorera et al., 2016).
Beside data on clinically relevant ATB resistance in surface waters, the c-IC determination provides critical information on the ecosystem vulnerability. The increase in ATB concentrations leads to a decrease in bacterial abundance, but the latter is not systematically linked to a decrease in the taxonomic richness, as observed in the Mauguio Lagoon samples. Resistant communities in the coastal lagoon waters present a singular behavior under ATB exposure, which differs from that of the resistant communities in the coastal river flowing to the lagoon. Indeed, the overall antimicrobial resistance of the brackish-water community is combined with a rather stable community structure. Most genera are probably autochthonous of the lagoon ecosystem (Clementino et al., 2008; Esteves et al., 2015), including Pseudomonas and Vibrio, which are potentially pathogenic for human and other animals. This relatively low vulnerability to ATB exposure suggests that the Mauguio Lagoon is probably a stable reservoir for resistant bacteria and resistance genes. The fact that low ATB concentrations decrease the abundance but not the taxonomic richness of the lagoon waters is rather paradoxical. One hypothesis is that every OTU identified up to the genus level corresponds to a heterogeneous population, grouping variants that display diverse behaviors against antimicrobial agents. Such diversity confers an “insurance effect” to the population by increasing the range of conditions in which the community as a whole can thrive (Boles et al., 2004). In contrast, for freshwaters from the Lez River and the Salagou Lake, various selective pressures (ATB types and concentrations) jumble the bacterial community structures. The changes observed consist in the selection of dominant OTUs, which belong to genera known to include human pathogens like Pseudomonas, Enterobacteriaceae, Acinetobacter, and Aeromonas. Despite the Lez River and the Salagou Lake differ in term of anthropogenic pressure, the quantitative and qualitative resistances of their bacterial communities are roughly similar. Surprisingly, ATB-resistant and potentially pathogenic Enterobacteriaceae and Acinetobacter are mostly selected in the lake water, which is less anthropized than the Lez River water.
5 Conclusion and perspectives
New concepts of resistome and pathobiome (D’Costa et al., 2006; Perry et al., 2014; Romano-Bertrand et al., 2015; Vayssier-Taussat et al., 2014) show that microbial communities in diverse aquatic environments can act as “incubators” of resistant bacteria that can be later involved in human infectious diseases. Environmental microbiology studies would benefit from the evaluation of ATB resistance of bacteria by c-IC determination in order to: (i) characterize bacterial communities and compare their overall profile in terms of antimicrobial resistance, (ii) evaluate the vulnerability of aquatic ecosystems under pressure of antimicrobial agents, (iii) characterize clinically resistant members among these ecosystems, and (iv) study the influence of local pressures on the resistome design and dynamics. Finally, c-IC determination can be easily adapted to monitor specific emerging resistances in the environment, such as the worrisome resistance to carbapenems (Woodford et al., 2014).
Acknowledgements
This work was supported by funding provided by the Human-Environment Observatory of the Mediterranean Coast (OHM “Littoral méditerranéen”), the DRIIHM Labex, CNRS, and by the “Programme national EC2CO Écosphère continentale et côtière”, CNRS, and by a doctoral fellowship (Ayad Almakki) from Campus France. We thank Dr. H. Jean-Pierre for fructuous discussions on resistance to ATB in medicine.


