1 Introduction
Electromyography (EMG) covers a set of electrophysiological techniques allowing diagnosing neuromuscular diseases. The diagnostic procedure involves the systematic acquisition of numeric and symbolic data; it proceeds according to a cycling set of steps–formulation of hypotheses, launching of dedicated examination procedures, evaluation of results, validation or questioning of the current hypothesis–until the final diagnosis formulation. The domain is broad, covering more than 100 diagnoses, and including 1000 tests of nerve or muscle structures.
To cope with the complexity of EMG diagnosis, several computerised systems have been developed to support the collection of data, facilitate their analysis and assist their interpretation. Various knowledge-based systems have been developed, among which SESAME 〚1〛 KANDID 〚2〛, ADELE 〚3〛, NEUROP 〚4〛, NEUROMYOSIS 〚5〛 and HINT 〚6〛. Causal functional model has been used in DARE 〚7〛, whilst Bayesian approaches have been considered in MUNIN 〚8〛 and DIAGNOSTICA 〚9〛.
Due to the recent development of the Internet technology, there is nowadays a growing demand for disseminating and sharing the medical expertise in this field, for building consensual databases and for sharing tools for their exploitation 〚10〛. This is the major goal of the EMG-Net project, which was conducted under the EEC Inco-Copernicus framework. The guiding principle of our work, more precisely, has been to design a web-based system integrating knowledge and data management facilities, with the aim of benefiting from a coupling between knowledge-based representation of medical expertise and data-centred mining of the available cases.
Data mining nowadays encompasses a large number of tools and methods, stepping from pure statistics to learning and visualisation: the accuracy and interpretability of this step are known to be key features in medicine 〚11〛. It implies considering data mining as a complex process whose main features are (i) to be grounded in medical knowledge, (ii) to support multistrategy methodologies and (iii) to be carefully controlled: combining knowledge-centred modelling with agent-centred design is advocated in this paper as a way to cope with the above-mentioned issues.
The background of our work is first of all briefly stated, before describing the knowledge management platform that forms the basis of our design; an agent-centred approach for data exploitation is then presented, together with some preliminary results in information retrieval and data mining.
2 Background
Data mining appears central to the development of advanced computer-aided decision-making systems in the field of medicine, by allowing the acquisition of a sound statistical knowledge, which, used in combination with the medical expertise, is the key to develop an evidence-based decision-making approach. The following issues are discussed as key milestones in this respect.
2.1 Accuracy and interpretability
EMG data are sparse, voluminous, multimodal and time-dependent; they are poorly reproducible, due to variations in the acquisition process, but also at the individual level; they are incomplete, due to possible variations in the diagnostic process that may influence the investigation protocol.
The data mining process in itself is a complex process to be driven carefully: the accuracy of computation must be asserted carefully, and its relevance must be checked. A multistrategy methodology has to be designed to cope with the wide variety of tools and approaches that form the core of knowledge discovery 〚12〛. Each one of these tools is known to play a specific role with respect to the whole data mining process, and moreover works under specific conditions, with a given focus.
To be usable, in addition, an advanced mining system should assist the user in the formulation of system-dependent goals: specific attention has to be paid in this respect, due to the necessity to handle statistical concepts that may not be familiar to the physician. Finding knowledge models able to integrate the statistical and medical knowledge is a key issue in this respect; grounding the data mining process into the domain ontology moreover appears essential to provide the user with domain-dependent query language.
The ability to bridge the gap between data (e.g. casual elements stored into tables, under raw format) and knowledge (e.g. generic information commonly stored into objects, under frame-like format) is consequently crucial: the growing usage of object-centred modelling brings a step in this direction, and there is a constant need to improve the quality of data representation and modelling in order to exploit and learn from them more easily. As a matter of fact, object-oriented data models embody rich data structures and semantics that appear highly beneficial to the knowledge discovery process 〚13〛.
Nowadays, data mining is increasingly considered as a cooperative process relying on strong knowledge and data structuring, knowledge being used to drive and assist the analysis and interpretation phases, these phases in turn bringing new unforeseen knowledge elements 〚14〛. Conversely, being rooted into the domain ontology, data mining results are stored under a format more easily understandable for the user.
2.2 Agent-centred approaches to data mining
An agent-centred design is advocated in this paper as a way to cope with the above-mentioned issues.
The algorithms that are commonly used during the data mining process may be rather complicated to operate due to the heterogeneity of data types and locations: the data to analyse is very often distributed among several databases, where it may be stored under different formats. To deal with such issues, multi-agent design is increasingly advocated. The principle is to distribute the mining process to several agents that have the capacity to communicate by sending messages. This distribution allows to access distributed databases in parallel 〚15, 16〛. So-called ‘configuration’ or ‘facilitator’ agents are used in this case (i) to provide registration services to member data sites, (ii) to coordinate their activity and (iii) join their results.
Moreover, agents are often designed as autonomous entities, which can learn from their environment, from the user and from other agents; as a consequence, they may adapt dynamically to the user profile and problem features; in addition, being autonomous entities, they may create other agents working under new goals, to complete, validate or discover some information 〚17〛. Active mining for example has developed recently 〚18〛 as performed by autonomous mining agents able to seek out interesting subsets and to ‘fly’ to them. Such agents are meant to operate autonomously but in a way that is grounded in the knowledge of the application domain and user’s profile, thus performing customised search.
Such collaborative perspective nowadays knows a growing interest among the Information Technology (IT) community and decentralised; simultaneous searching in several distributed databases is already possible. Collaborative approaches may also be used to assist agents in the development of the search process, or in the learning of customised behaviours 〚19〛.
3 Description of the platform
The platform architecture, which is presented in this paper, follows a client–server approach that could provide specific client user interface with common servers.
The platform inherits from ESTEEM Project AIM No. 2010 and EMG-Net Project INCO No. 979069. These projects provided a significant amount of knowledge and data that have been partially standardised. This information is currently stored within the EMG databases. In order to go further into EMG standardisation, and to complete and enhance the EMG knowledge, the proposed platform has been designed to involve four main modules (Fig. 1):

The EMG platform allowing access to knowledge base, data-mining modules and EMG database. The server of the platform 〚20〛 is implemented under free portable software AROM 〚21〛. AROM (http://www.inrialpes.fr/sherpa/pub/arom/) has been provided with a web interface, WebAROM, which allows sharing and exploit knowledge from everywhere through the Internet networks in a friendly way.
- • the EMG Data-base to store the patient cases,
- • the data-mining modules to gain knowledge about the medical domain,
- • the knowledge-base module to store the knowledge partially standardised,
- • the worldwide server to disseminate the results and interact with the user in a friendly way.
The patient database is developed through a general data structure covering all anatomical structures, examination techniques and parameters used by the physicians of the two European projects (ESTEEM and EMG-Net). The data structure has been implemented in several computer programs allowing: (1) the transfer of data from different EMG machines, (2) the local sampling and interpretation of EMG examinations, (3) the exchange of EMG examinations between laboratories and (4) the transfer of EMG examinations to and from different decision support systems (DSSs). This database is implemented under SQL architecture.
The EMG knowledge base formalises the EMG expert knowledge: this means that the expert knowledge is expressed in a formal language carrying a precise semantics. The knowledge base is built in AROM language, and defines the elements that the different data mining modules will be able to operate on.
AROM provides a framework in the form of top-level declarative abstraction hierarchies, in which the EMG expert practitioners create the consensual EMG knowledge base. These top-level abstraction hierarchies are represented with enough information for modelling the EMG domain. The representation model articulates the two notions of classes and associations, the latter notion being used to represent and describe various types of relations between classes. In addition, AROM is provided with an Algebraic Modelling Language (AML) 〚22〛, which provides a formal support for the expression of constraints or queries. Thanks to the presence of the AML, this system is more particularly adapted to application domains combining both numeric and symbolic information, and its extension to integrating data and knowledge management looks quite natural.
4 An agent-centred design for data management in EMG
In the following, we present an agent-centred framework whose targeted goal is (i) to assist the retrieval of information about cases and (ii) to discover associations between data items, based on some knowledge-driven user formulated request.
4.1 Designing approach
Information retrieval and data mining tools have been added to WebAROM under the JSP–Java Server Pages–technology (a technology allowing the creation of dynamic html pages from Java programs). A nice coupling between knowledge-centred and data-driven analysis is obtained as a consequence, since the AROM knowledge model is used to assist request formulation and to drive the data exploitation process; data exploitation in turn is considered as a way to potentially enrich the knowledge base.
The interest of such approach is twofold: data mining is grounded into the domain knowledge, thus bridging the gap between statistical knowledge and medical expertise 〚23〛; as a consequence, request formulation is approached in terms familiar to the physician 〚24, 25〛.
SQL (Standard Query Language) is currently used as data modelling language. The databases are organized around three main information levels:
- • examination level (e.g. age of the patient, laboratory of examination, final diagnosis...);
- • anatomical level (e.g. examined structure types, conclusion about these structures...);
- • test level (e.g. type of test performed for a given structure, result for this test, type of test conclusion...).
4.2 Agent roles
Dedicated agents have been designed to support the information retrieval and data mining activities; as depicted in Fig. 2, these agents are of three basic types:

Agent roles and coordination.
- • the task-agent plays the role of an interface between the user and the retrieval/mining processes; as such, its role is twofold: it is (i) to receive the user query and transmit the results back to the user, and (ii) to translate the query in terms of sub-tasks to be performed by sub-agents and to recover their results; this agent plays the role of the ‘facilitator’ agent advocated in the introduction part;
- • the sub-agent is coupled with a specific type of information, e.g. information related to the anatomical structures or to the patient examination; its role is (i) to manage the part of a query addressing a given type of information and (ii) to distribute this query, which may imply several kinds of information for a given type, to corresponding data-agents;
- • each data-agent is coupled with a specific table in the database; its role is to process a ‘simple’ request, i.e. a request involving one parameter and one field of information.
As may be seen from Fig. 2, the data management system is organised into successive layers, which ensure the circulation of information, and its progressive transformation, from data to knowledge and vice-versa, under dedicated agent control. As a consequence, each agent is meant to work in a rather focalised way, under specific goals and assumptions.
Each agent is implemented as a Java object and communicates with the databases by means of JDBC (Java DataBase Connection). The agents communicate through message sending and execute under control of an administrator as an endless message waiting cycle. The mining resource at their disposal is the well-known association rule computation 〚26〛. Such rules show attribute-value conditions that occur frequently together in a given set of data: an association rule of the form X → Y is interpreted as “database tuples that satisfy X are likely to satisfy Y”.
Such design allows approaching the data management task under a cooperative assumption, the task being distributed to specialised processing entities. Better control is moreover exercised, and allows for example coping with data incompleteness: each data agent has full local control over the information support needed to answer a request; partial replies may be issued at the sub- and task-agent level, when necessary.
5 Information retrieval
The aim of the information retrieval functionality is to retrieve the diagnostic information corresponding to some user specified parameters. An example request is “to retrieve the diagnoses done for heredity reasons on male patients, with a neuropathic structure conclusion concerning the medianus nerve”. The corresponding repartition of roles and processes, at the agent level, is provided in Fig. 3 for the above-mentioned example request. The request is distributed to sub-agents and then to data-agents that collect the corresponding information in a distributed way. The results are then joined successively at the sub-agent and task-agent levels before transmission to the requesting user.

Information retrieval: functional pattern example.
The information retrieval interface is shown in Fig. 4. Several windows allow the user to browse easily among the various types of information and to select accordingly the research criteria. As may be seen from this figure, this process takes place in the framework of the knowledge ontology supported by AROM.

A view of the information retrieval interface.
Information retrieval results are provided as depicted in Fig. 5. The list of EMG diagnoses corresponding to the user request is displayed together with their frequency of appearance, and their confidence values, as attributed by the physician during the examination and recorded in the database. The diagnoses are sorted according to their frequency of appearance.

Information retrieval results: an example view.
Finally, the system further provides, for each diagnosis, the possibility to go back to the effective data corresponding to any retrieved examination. Such ‘feedback’ allows the user to check rapidly, under a synthetic presentation, the information shared or not by examinations verifying a common request. A further functionality, which is currently under development, is to launch a PCA analysis on the retrieved information, allowing examining the distribution of information, in terms of a user selected qualifier.
6 Data mining
The data mining functionality is currently implemented as a mere association rule computation 〚26〛. The system may for example look for associations between two nerve structures, e.g. Tibialis and Peroneus nerves.
The process implementation is exemplified in Fig. 6: the request is first of all distributed to the corresponding data tables, the results being joined in a second step.
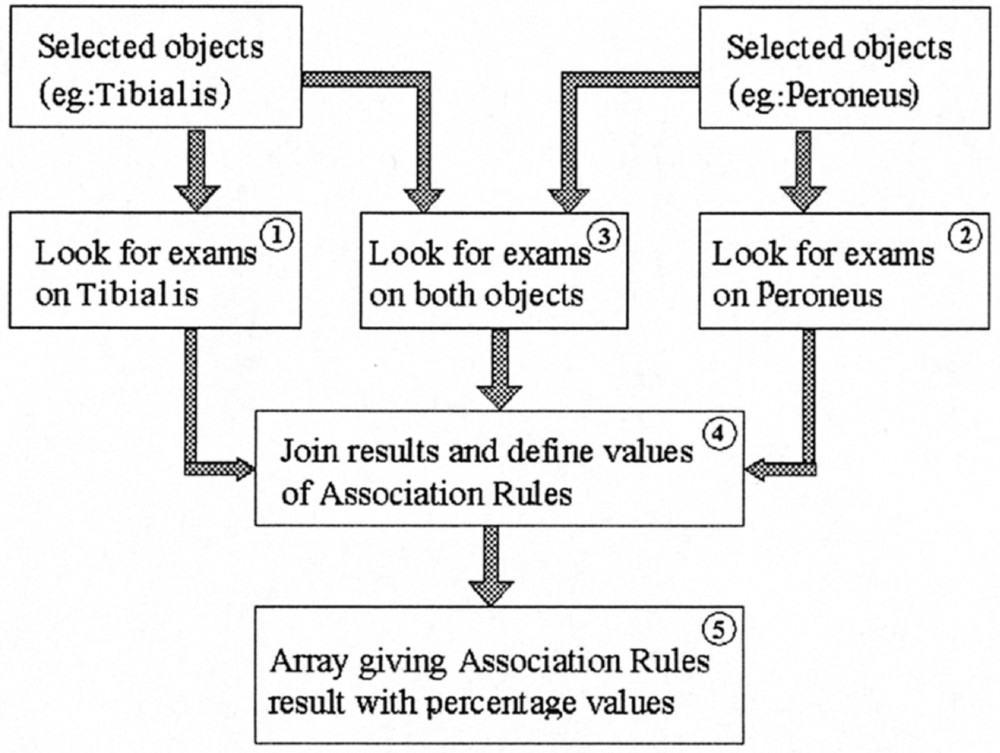
Data mining process: functional pattern example.
The request is formulated under the WebAROM interface, as shown in Fig. 7, under a two-step process: an item of interest is selected first; the second item is then selected, in the context of the previous one.

Association rules description.
The result of a given search is displayed in a dedicated window, as shown in Fig. 8.
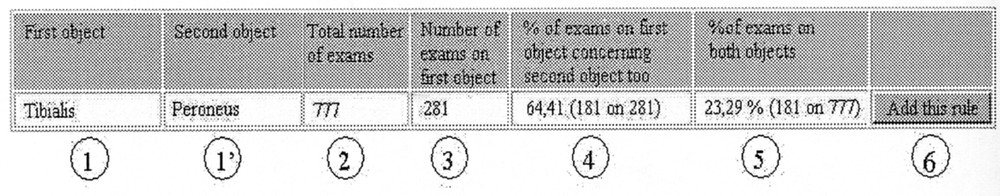
Association rules functional pattern.
The button ‘add this rule’ finally allows to add the new association rule in the knowledge server and thus to enrich and complete the knowledge base with new concepts (Fig. 9).

Adding a new association rule into the AROM knowledge base.
7 Conclusion and perspectives
The use of agent-centred design has been advocated in this paper as a way to cope with the specificities of data management in the medical domain, and more specifically in EMG, and a rough sketch of an agent-centred platform that is currently under development in the framework of EMG has been presented.
Information retrieval as well as data mining processes has been designed at the interface between data and knowledge, thus rendering the system usage more easy to the physician but also considering the extraction of knowledge as the ultimate goal of data exploitation.
An agent-centred approach has been considered to ensure processing efficiency, in front of potentially large amount of data. Another advantage of a multi-agent approach is to allow distributing the work among autonomous entities able to communicate, cooperate and learn from one another.
Further development implies increasing the agent communication and cooperation abilities as well as increasing the data management resources at hand. Using the AML language is currently under study to enrich query formulation.
Extensive discussion with the end-users will finally be necessary to delimitate the system potential and ensure its effective usage.
Acknowledgements
This work has been conducted under the auspice of the EEC INCO-COPERNICUS program. European Project EMG-net No. 977069.
Version abrégée
Le travail décrit dans cet article a été conduit dans le cadre d’un projet Inco–Copernicus, dont l’objectif était de concevoir un système accessible via le web et offrant des facilités de gestion des données et des connaissances en électromyographie. Ce domaine d’application est en effet très vaste ; il recouvre plus de 100 diagnostics différents et implique plus d’un millier de tests des structures nerveuses et musculaires. Un nombre significatif de cas a pu être collecté lors de projets précédents et stocké dans des bases de données. Des modèles de la connaissance médicale et du protocole d’analyse ont également été élaborés.
Notre objectif était de tirer parti de l’ensemble de ces données et connaissances, jusque-là spécifiées séparément, en mettant en œuvre des processus de fouille de données, dirigés par les connaissances du domaine.
Une plate-forme a été conçue dans ce but, dotée d’une architecture client–serveur. Elle comprend plusieurs modules :
- • la base de données ;
- • la base de connaissance ;
- • les modules de fouille de données ;
- • le serveur web.
La base de données est implantée selon le modèle relationnel ; elle est organisée en trois niveaux : niveau de l’examen (données patient, laboratoire et examinateur, diagnostic final, …), niveau de l’anatomie (type des structures examinées, conclusions sur ces structures, …) et niveau des tests (type de test effectué sur les structures, résultats des tests...).
La base de connaissances est implantée sous Arom, un langage de représentation des connaissances développé en Java exploitant les deux notions de classes et d’associations. Arom est doté, en outre, d’un langage algébrique, qui offre un cadre formel pour l’expression de contraintes ou de requêtes.
Les modules de fouille de données (calcul de règles d’associations) sont appelés par des agents exploitant la base de données de manière concurrente. Des agents de type « tâche » interprètent la requête et la distribuent à des agents de niveau inférieur, chargés de traiter la requête, de collecter les résultats et de les transmettre au niveau supérieur. L’information transite ainsi depuis le niveau des connaissances (formulation de la requête) jusqu’au niveau des données (traitement de la requête) ; en outre, les agents travaillent de manière focalisée et coopérative, selon des buts précis. Les agents sont implantés en Java et communiquent avec les bases de données via JDBC (Java DataBase Connection). Ils communiquent entre eux par envoi de messages et s’exécutent sous le contrôle d’un administrateur, selon un cycle d’attente infinie de messages.
Le serveur web permet l’exploitation des données à distance, selon un style d’interface maintenant familier à beaucoup d’usagers.
La plate-forme offre actuellement deux formes de services : recherche d’information et fouille de données.
La fonction de recherche d’information permet de collecter les informations diagnostiques correspondant à des cas dont les caractéristiques sont spécifiées par l’utilisateur. Une requête est par exemple de la forme : « trouver les diagnostics émis pour des raisons d’hérédité sur des patients de sexe masculin, et pour lesquels une neuropathie a été décelée sur le nerf médianus ». Une telle requête est formulée aisément, à l’aide d’une interface permettant de naviguer au sein des concepts et attributs du domaine. Il est ensuite possible de revenir en détail sur les informations relatives aux cas extraits.
La fonction de fouille de données permet la recherche de règles d’association entre les données. Ces règles, de la forme « condition–conclusion », représentent des associations trouvées fréquemment dans les données ; une règle de la forme X → Y s’interprète ainsi comme « une entité de la base de donnée qui satisfait X satisfait probablement Y ». On cherchera par exemple la fréquence de l’association « nerf Tibialis » et « nerf Peroneus » au sein de la base de cas, une fréquence élevée indiquant que les deux examens sont fréquemment associés.


