1 Introduction
From various perspectives, the Praomys group is one of the most successful groups of Old World rodents. Taxonomically, their diversification is important, and one may conservatively include in it, as a starting point, six genera (Heimyscus, Hylomyscus, Mastomys, Myomys, Praomys and Stenocephalemys), comprising 34 currently recognised species 〚1, and references hereunder〛. Ecologically, they have colonised various biotopes, ranging from the equatorial rain forest to Sahelian savannas. At the community level, many of them can build abundant populations. Finally, their interactions with human populations can have important consequences with regard to agriculture and human health. The systematics of the whole group, as well as its constituent taxa, has been very unstable, experiencing many changes according to taxonomic treatments and authors 〚2–10〛. At various times, different authors have either included the current genera as sub-genera within Praomys, or have suggested linkages between them. For example, Hylomyscus, Mastomys and Myomys were considered as sub-genera within Rattus/Epimys or Praomys 〚2, 11, 12〛. Heimyscus fumosus was originally considered to be a member of Hylomyscus 〚13〛, while Van der Straeten and Dieterlen 〚14〛 originally described Myomys ruppi as a species of Praomys, sharing features with Myomys albipes and Stenocephalemys. Moreover, different authors have included additional genera (Zelotomys 〚10, 15〛, Stenocephalemys 〚14, 16, 17〛, Colomys 〚16〛, Heimyscus 〚4, 13〛) or species (Malacomys verschureni 〚18〛), within the Praomys group. Here, we refer to the ‘Praomys group’ as this extended suite of genera and species. However, the relationships between and within these taxa are still not well understood. This is mainly due to the low level of morphological differentiation among the species as well as between the genera, making it difficult to find phylogenetically informative characters. Further, the monophyly of the genera Praomys, Myomys, and Stenocephalemys has been questioned by several studies 〚16, 17, 19, 20〛. Up to now, the majority of studies on the systematics of the Praomys group have been based on phenetic analyses (morphological, biometrical or molecular) and have involved relatively few of the relevant taxa. Only one study, based on a cladistic analysis of morphological characters on most of the African members of the Praomys group is available to date 〚20〛.
Here we present the first cytochrome b-based phylogeny of the Praomys group as represented by a significant sample of species from all currently recognised genera. More precisely, this study is the first where almost all the Myomys species are considered, as well as the species Mastomys verheyeni and Mastomys pernanus. It also includes many species from the other genera comprising the Praomys group, including the poorly known Zelotomys hildegardeae, Colomys goslingi and Malacomys verschureni. The monophyly of these genera will be tested as well as the various hypotheses concerning their relationships. Taxonomical and nomenclatural implications will be proposed and discussed. These results provide insights into the chronological succession of divergence events in this group. Based on divergence time estimates, we discuss some of the possible patterns of evolution among the Praomys group.
2 Material and methods
2.1 Species sampling
A total of 58 specimens of murine rodents representing 36 different taxa were included in the phylogenetic analyses. In the Praomys group, the genus Praomys was represented by P. tullbergi, P. misonnei, P. sp nov 〚21〛, P. hartwigi obscurus, P. jacksoni and P. degraaffi; Mastomys by M. natalensis, M. erythroleucus, M. coucha, M. huberti, M. verheyeni and M. pernanus; Myomys by M. albipes, M. daltoni, M. derooi, M. fumatus, M. verreauxii and M. yemeni; Hylomyscus by H. stella, H. parvus and H. alleni; Heimyscus by H. fumosus; Stenocephalemys by S. albocaudata and S. griseicauda. Colomys goslingi, Zelotomys hildegardeae and Malacomys verschureni were also studied. For the list of material see Table 1 . Some taxa were included from Genbank: Mastomys huberti (AF141220) and the outgroup taxa Arvicanthis niloticus (AF004572), Hybomys univittatus (AF141219), Lemniscomys bellieri (AF004586), Dasymys incomtus (AF141217), Aethomys sp (AF004587), Rattus norvegicus (AB033713), Mus musculus (AB033699), Apodemus flavicollis (AB032853) and Apodemus sylvaticus (AB033695), which were chosen following the results of previous molecular analyses 〚6, 22〛.
List of the specimens examined with geographic origin, collector and collection number. MNHN: Muséum national d’histoire naturelle, Paris; CM: Carnegie Museum, Pittsburgh; FNS: Forschungsinstitut und Naturmuseum Senckenberg, Frankfurt; RUCA: Department of Biology of the University of Antwerp; FMNH: Field Museum of Natural History, Chicago; SMNS: Staatliches Museum für Naturkunde, Stuttgart; ZFMK: Zoologisches Forschungsinstitut und Museum Alexander Koenig, Bonn; T numbers refer to the catalogue of mammalian tissues curated at the University of Montpellier.
| Specimens | Geographic origin | Collectors | Museum number |
| Hylomyscus alleni | Gabon, province estuaire | LW. Robbins | CM - TK 21550 |
| Hylomyscus parvus | Cameroon, Ikenge | DA. Schlitter | CM - SP 10502 |
| Hylomyscus parvus | Cameroon, Ikenge | DA. Schlitter | CM - SP 10514 |
| Hylomyscus stella | Kenya, Kakamega | DA. Schlitter | CM - SP 5032 |
| Heimyscus fumosus | Gabon, R. de la Moukalaba | V. Nicolas/E. Lecompte | MNHN - 2001-064 |
| Heimyscus fumosus | Gabon, R. de la Moukalaba | V. Nicolas/E. Lecompte | MNHN - 2001-076 |
| Mastomys coucha | South Africa, Bloemfontein | P. Taylor | MNHN - 1999-104 |
| Mastomys coucha | South Africa, Bloemfontein | P. Taylor | MNHN - 1999-106 |
| Mastomys erythroleucus | Mali, Kangaba | B. Sicard | MNHN - 1999-322 |
| Mastomys erythroleucus | Ethiopia, Mizzan-Tefferi | L. Lavrenchenko | MNHN - 1999-551 |
| Mastomys erythroleucus | Chad, Tinga | C. Denys/L. Granjon | 4123 |
| Mastomys huberti | Mali, Emnal’here | B. Sicard | SK 317 |
| Mastomys natalensis | Senegal, Kédougou | K. Bâ | MNHN - 1999-349 |
| Mastomys natalensis | South Africa, Badplaas | P. Taylor | MNHN - 1999-099 |
| Mastomys natalensis | Zimbabwe | P. Taylor | MNHN - 1999-110 |
| Mastomys pernanus | Tanzania, Dakawa | H. Leirs | RUCA - 1624 |
| Mastomys verheyeni | Nigeria, Dugumne | W. Brei | SM - 54752 |
| Mastomys verheyeni | Nigeria, Dugumne | W. Brei | SM - 54755 |
| Myomys albipes | Ethiopia, Mizan-Tefferi | L. Lavrenchenko | MNHN - 1999-561 |
| Myomys albipes | Ethiopia, Entoto | M. Corti | T-1581 |
| Myomys daltoni | Mali, Gao | B. Sicard | MNHN - 1999-392 |
| Myomys daltoni | Benin, Deroubanou | W. Brei | SMNS - 39353 |
| Myomys derooi | Ghana, Yendi | DA. Schlitter | CM - SP 10190 |
| Myomys derooi | Benin, Partago | W. Brei | SMNS - 39394 |
| Myomys fumatus | Kenya, Naivasha | DA. Schlitter | CM - TK 33059 |
| Myomys fumatus | Kenya, Naivasha | DA. Schlitter | CM - TK 33060 |
| Myomys verreauxii | South Africa, Paarl Moutains | P. Taylor | MNHN - 1999-102 |
| Myomys verreauxii | South Africa, Cape | P. Taylor | MNHN - 1999-112 |
| Myomys yemeni | Saudi Arabia, 20 km Abha, Taïf road | P. Gaucher | MNHN - 1994-079 |
| Myomys yemeni | Saudi Arabia, Alba | P. Gaucher | T-1457 |
| Praomys degraaffi | Burundi, Kibira National Park | JC. Kerbis | FMNH - 138046 |
| Praomys degraaffi | Uganda, Gahinga gorilla NP | JC. Kerbis | FMNH - 157790 |
| Praomys jacksoni | Gabon, Mayibout | C. Fournier-Chambrillon | MNHN - 1999-402 |
| Praomys jacksoni | Uganda, Kalanga | JC. Kerbis | FMNH - 138095 |
| Praomys misonnei | Uganda, Budongo | JM. Chupasko | FMNH - 165317 |
| Praomys misonnei | Kenya, Kakamega | DA. Schlitter | CM - SP 5018 |
| Praomys misonnei | Democratic Republic of Congo, Epulu | JC. Kerbis | FMNH - 149594 |
| Praomys tullbergi | Ghana, Katamanso | DA. Schlitter | CM - SP 10188 |
| Praomys hartwigi obscurus | Nigeria, Gangirwal | G. Nikolaus | SMNS - 41313 |
| Praomys hartwigi obscurus | Nigeria | G. Nikolaus | ZFMK - 88-130 |
| Praomys sp. nov. | Congo, Tchissanga | L. Granjon | T-795 |
| Stenocephalemys albocaudata | Ethiopia, Sanetti | M. Corti | T-1590 |
| Stenocephalemys albocaudata | Ethiopia, Sanetti | M. Corti | T-1589 |
| Stenocephalemys griseicauda | Ethiopia, Mt Chilalo | H. Rupp | SMNS - 35945 |
| Colomys goslingi | Sudan, Gilo | G. Nikolaus | SMNS - 30124 |
| Malacomys verschureni | Democratic Republic of Congo, Irangi | F. Dieterlen | SMNS - 131 |
| Zelotomys hildegardeae | Uganda, Queen Elizabeth Park | A. Hoffman | 3190 |
| Zelotomys hildegardeae | Uganda, Queen Elizabeth Park | A. Hoffman | 3191 |
2.2 DNA extraction, amplification and sequencing
Total genomic DNA was extracted from liver, heart or muscles preserved in 70% ethanol using a CTAB protocol 〚23〛. Mitochondrial sequences containing the cytochrome b gene were isolated via the polymerase chain reaction (PCR). The entire cytochrome b gene was amplified using the primers L14723 (5’ ACC AAT GAC ATG AAA AAT CAT CGT T 3’) and H15915 (5’ TCT CCA TTT CTG GTT TAC AAG AC 3’) in the flanking region of the cytochrome b gene. Additional internal primers, used when the DNA was too fragmented and for the sequencing, were H15553 (5’ TAG GCA AAT AGG AAA TAT CAT TCT GGT 3’), L15408 (5’ ATA GAC AAA ATC CCA TTC CA 3’), L15146 (5’ CAT GAG GAC AAA TAT CAT TCT GAG 3’), H15149 (5’ CTC AGA ATG ATA TTT GTC CTC 3’), L15513 (5’ CTA GGA GAC CCA GAC AAC TA 3’). Primers name indicates the DNA strand (H, heavy or L light) and the position of the 3’ end of the oligonucleotide.
Double-stranded PCR amplifications were performed in 50 μl reaction volumes using primers L14723 and H15915. Each reaction included 0.6 μl primers (50 pM μl–1), 2.5 μl of DMSO, 2 μl of desoxynucleoside–triphosphate mixture, 5 μl of reaction buffer (10X) (Appligen) and 0.3 μl of taq DNA polymerase (Appligen). All PCRs used the following thermal cycling parameters: 4 min at 94 °C, 36 cycles (40 s at 94 °C, 45 s at 50 °C, 40 s at 72 °C). Double-stranded PCR products were purified from agarose gel using the MinElute purification kit (Qiagen). Taxa were sequenced directly from purified PCR products using primers mentioned above. Approximately 100 pmol ml–1 of double-stranded PCR product was used in an automatic sequencer CEQ2000 (Beckman). The sequences were entered and manually aligned using the Bioedit software 〚24〛. All the sequences are available in Genbank under accession numbers AF518328–AF518375.
2.3 Saturation analyses
Mutational saturation was studied for each codon position, for transitions and transversions separately, by plotting the pairwise observed number of sequence differences (in percentage) against the pairwise number of substitutions in the maximum parsimony tree as inferred by PAUP 3.1.1. 〚25〛.
2.4 Phylogenetic analyses
Phylogenetic relationships were analysed by maximum-parsimony (MP), neighbour-joining (NJ) and maximum-likelihood (ML) methods. The phylogenetic analyses were conducted using PAUP 4.0b4a 〚26〛. Critical values of skewness (g1 statistics) of the tree length distribution were used to access overall phylogenetic signal in the data set. The critical values were computed from a distribution of 10 000 randomly generated trees using PAUP.
The MP analysis was done with a heuristic search using stepwise addition. Robustness of trees was assessed by the bootstrap method 〚27〛 performed by PAUP (1000 replicates) and by the Decay Index (DI) 〚28〛 using the Autodecay software 〚29〛. The program Modeltest 3.04 〚30〛 was used to select the substitution model that best fitted the data according to hierarchical likelihood ratio tests. The NJ tree was performed from the distance matrix calculated under the selected model, and the robustness of inferences was assessed through bootstrap resampling (1000 replicates). The ML analyses was performed with ten random additions of taxa under the selected model, the robustness of inferences was assessed through bootstrap resampling (100 replicates) using PAUP 〚26〛. Analyses were also performed on the amino acid (AA) sequences, both with NJ and MP methods.
2.5 Relative-rate test
Relative-rate tests were conducted both with RRTree, version 1.0 〚31〛, by taking into account the taxonomic representativeness and its phylogenetic relationships, and with the Mega program 〚32〛. Quantifications of the rate differences were performed either on the proportions of synonymous (Ks) and non-synonymous (Ka) substitutions with RRTree, or on all three codon positions with Mega. Relative-rate tests were performed among the Praomys group species; Arvicanthis, Mus and Apodemus were chosen as outgroups.
3 Results
3.1 Cytochrome b gene sequences, base composition, saturation and sequence variation
Complete cytochrome b gene sequences were obtained for 48 specimens and analysed together with additional available Murinae sequences (cf. Table 1). In order to check for the mitochondrial origin of the cytochrome b gene sequences, the corresponding amino acid sequences were checked for the presence of aberrant amino acids (as stop codon), deletions or insertions. The sequence was 1140 bp long in all examined specimens, coding for 380 amino acids. The gene began in all studied specimens with the first three conserved codons (ATG, ACA and AAC) and there was no complete stop codon at the end of the gene, only a single T or C.
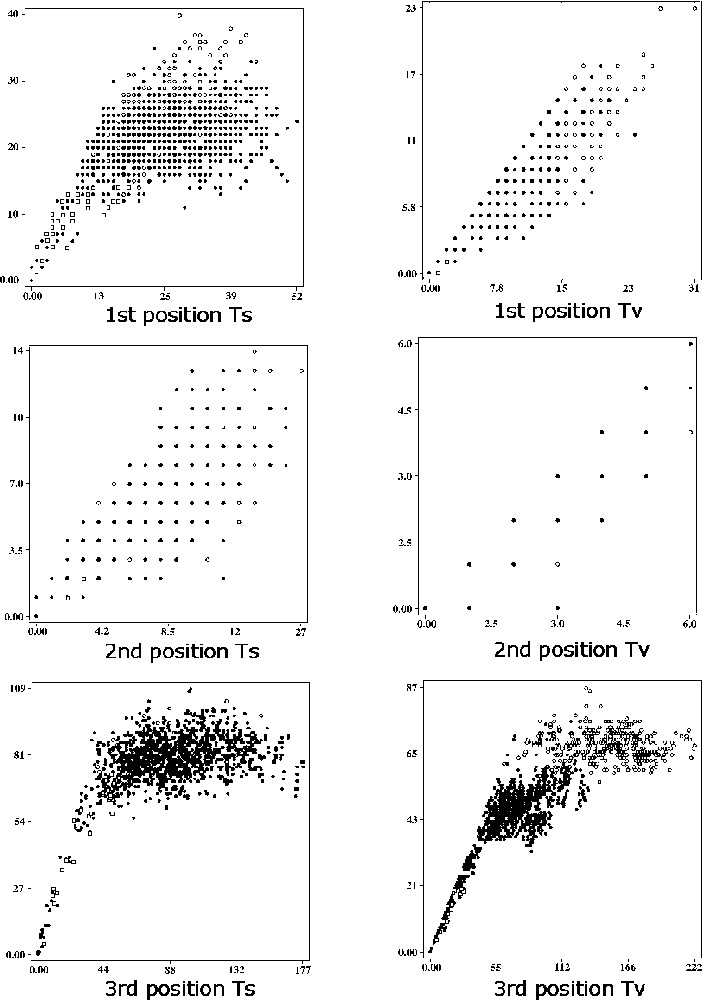
Graphs illustrating the levels of saturation in the complete 1140 pb data set at each codon position, for transitions (Ts) and transversions (Tv). Numbers of pairwise observed sequence differences (as a percentage, Y axis) are plotted against the corresponding numbers of pairwise inferred substitutions occurring along branches joining two species in the maximum parsimony tree of PAUP (X axis). The following symbols are used in all saturation graphs: full circles, comparisons within the Praomys group; open circles, comparisons of outgroup taxa with Praomys group; open squares, comparisons between Mastomys coucha, M. erythroleucus, M. huberti and M. natalensis.
Base composition of the cytochrome b gene in the examined Murinae was quite similar to that of previously reported mammalian sequences 〚33〛 and comparable to values found in some muroid rodents 〚34〛: the abundance of Gs was low (12.2%), whereas the percentages of A, T and C were quite similar (26.7–32.1%). As expected, the first and second positions showed less variability than the third codon positions 〚33〛.
The saturation curves are presented in Fig. 1. Transitions at the second codon position and transversions at the three codon positions were not saturated, while transitions at the first and third were saturated (Fig. 1). Consequently, the weighting scheme used has consisted in the elimination of transitions at the first and third position. Within the Mastomys species M. erythroleucus, M. natalensis, M. huberti and M. coucha, no saturation was found at any position (Fig. 1); so an independent analysis was conducted without any weighting for this group in MP and a ML analysis was conducted with a model selected by Modeltest 3.04 〚30〛 on the Mastomys dataset.
For the phylogenetic analyses, a data set of 58 specimens and 1140 nucleotides was used. These sequences could be unambiguously aligned. Of these sites, 563 were variable and 423 were parsimony informative; 140 (24.9%) of the variable nucleotides were at the first, 62 (11.0%) at the second and 361 (64.1%) at the third codon position. Of the phylogenetically informative sites, 84 (19.9%) were at the first, 30 (7.1%) at the second and 323 309 (73.0%) at the third codon position.
3.2 Genetic distances
Within the Praomys group, the inter-genera genetic distances ranged from 5.3% (between Myomys albipes and Stenocephalemys) to 17.9% (between Mastomys verheyeni and Mastomys pernanus). Within the genera, the range of divergence values was of the same order, going from 6.8% (within Stenocephalemys) to 17.9% (within Mastomys).
3.3 Phylogenetic analyses
The MP analysis on nucleotide data yielded 24 trees of 1033 steps, the strict consensus of which is presented in Fig. 2. The g1 statistics is significantly different (g1 = – 0.55, P < 0.01) from what would be expected from random phylogenetic noise. The NJ analysis on nucleotide data is presented in Fig. 3. The models selected were GTR + I + G for the whole Praomys group and TVM + I + G for the Mastomys data set. The ML analysis data yielded a tree highest-likelihood (ln L = 15 006.786), presented in Fig. 4. The MP analysis on amino acid data yielded 1886 trees of 436 steps (CI = 0.552); the MP and the NJ trees drawn from protein sequences (not shown) only display some groups already identified in the others analyses.
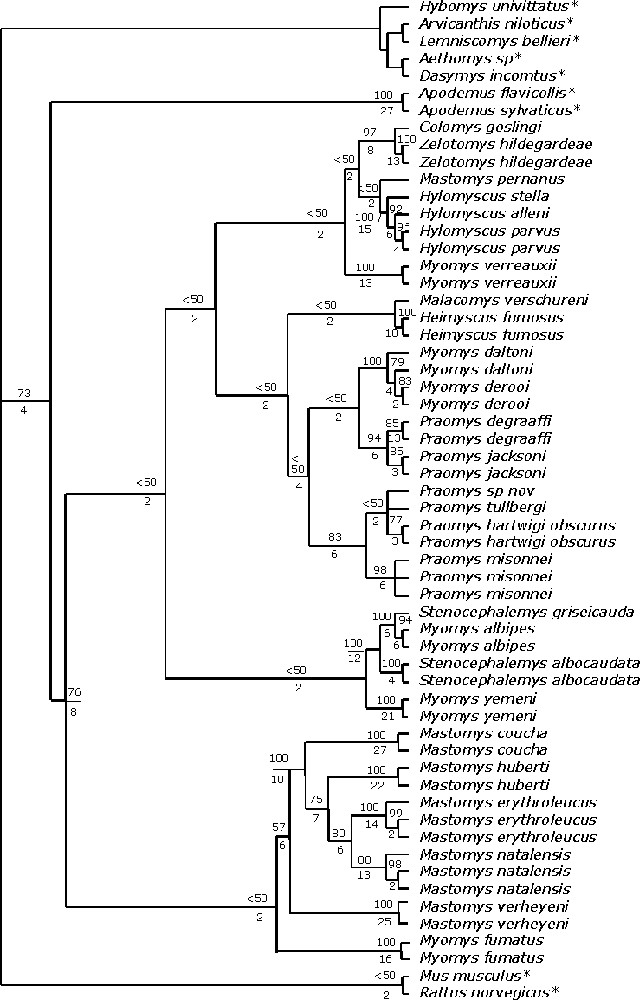
Phylogenetic relationships among nucleotide sequences of the complete cytochrome b gene (1140 pb) of 49 specimens of the Praomys group using maximum parsimony. The murid rodents used as outgroup are indicated with an asterisk. Trees are generated with a heuristic search (PAUP 4.0b4a) on non-saturated substitution (first and third position transitions removed). The numbers above branches are % bootstrap values (1000 replicates) and the number below represent the Bremer support Index. Strict consensus of 24 trees of 1033 steps, CI = 0.339, RI = 0.667, RC = 0.226.
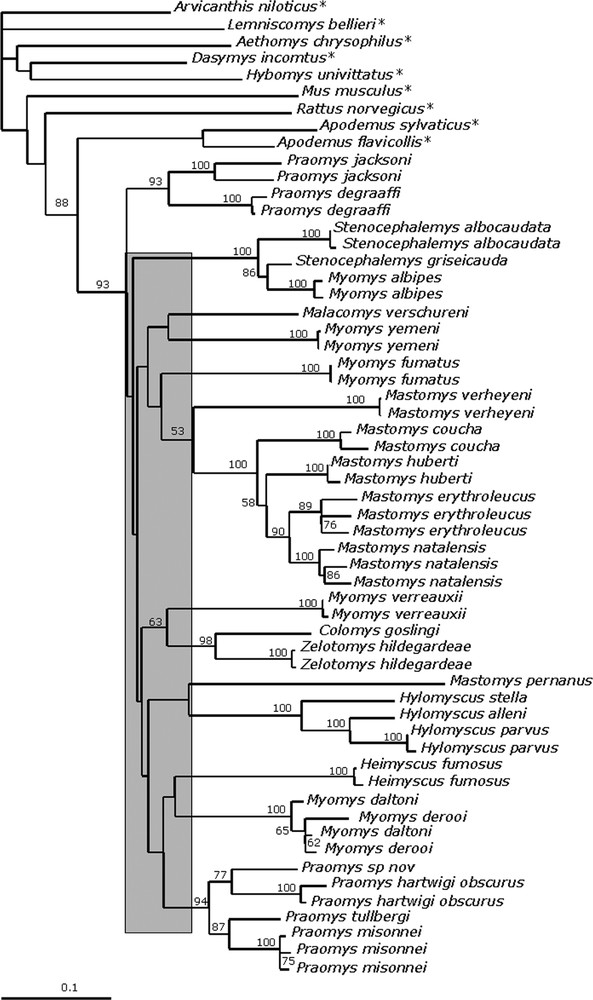
Phylogenetic relationships among nucleotide sequences of the complete cytochrome b gene (1140 pb) of 49 specimens of the Praomys group using the Neighbour-Joining method. The murid rodents used as outgroup are indicated with an asterisk. Tree was generated on distance matrix calculated under the model selected by Modeltest 3.04. The numbers above branches are bootstrap values above 50% (1000 replicates). Scale bar indicates 10% divergence. The grey area indicates the period of emergence of most of the major lineages.
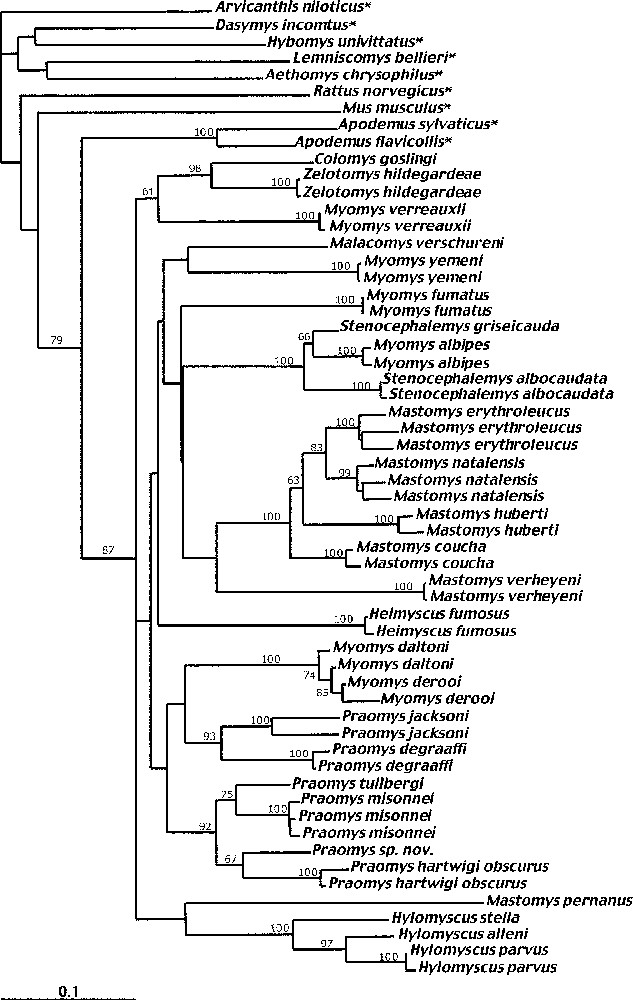
Phylogenetic relationships among nucleotide sequences of the complete cytochrome b gene (1140 pb) of 49 specimens of the Praomys group using maximum-likelihood method. The murid rodents used as outgroup are indicated with an asterisk. Trees are generated with an heuristic search (PAUP 4.0b4a), under the model GTR+I+G, selected by Modeltest 3.04. The numbers above branches are bootstrap values above 50% (100 replicates). Highest-likelihood tree (ln L=15 006.786). Scale bar indicates 10% divergence.
3.4 Phylogenetic relationships
With a few exceptions, MP, ML and NJ results are quite consistent with each other. Terminal clades are consistently confirmed in the different analyses and are typically well-supported in analyses based on DNA sequences. Some taxa or clades branching at the base of the Praomys group have an unstable position among the trees. The three ML, MP and NJ trees indicate that the Praomys group is monophyletic when it includes Malacomys verschureni, Colomys and Zelotomys (Figs. 2–4). This is supported, however, by quite a low bootstrap value of 76% in MP but with a high Decay Index value of 8 and a bootstrap value of 87% in ML and 93% in NJ. The sister group is the genus Apodemus (73% BP in MP, DI = 4; 88% in NJ and 79% in ML), both in analyses based on nucleotides and amino acids sequences.
The phylogenetic relationships among the genera within the Praomys group are not always clearly resolved, but some clades are strongly supported. For example, within the genus Praomys (which appears paraphyletic) two clades (jacksoni and tullbergi groups) are well supported (94% and 83% BP in MP; DI = 6; 93% and 92% in ML, respectively, Figs. 2 and 4). These two groups maintain their integrity in the NJ AA trees. The genus Myomys appears polyphyletic: Myomys fumatus clusters with Mastomys, while Myomys daltoni and Myomys derooi cluster with the genus Praomys. However, none of these phylogenetic relationships involving the Myomys species is strongly supported. The genus Stenocephalemys also appears to be paraphyletic. Along with Myomys albipes, Stenocephalemys, forms a strongly supported (100% BP in both MP and ML; DI = 12; Figs. 2 and 4) Ethiopian clade. This clade is also evident in the analyses based on amino acids. The monophyly of the genus Hylomyscus, represented here by three species, is strongly supported (100% BP in both MP and ML; DI = 15, Figs. 2 and 4) and is also recovered in the AA-based trees. Zelotomys hildegardeae appears as the sister group of Colomys goslingi with a high support (97% BP in MP; DI = 8; 98% BP in ML, Figs. 2 and 4), a relationship also found in the two AA-based analyses. Myomys verreauxii is found aligned to Colomys and Zelotomys, albeit with quite low bootstrap value (61% BP in ML; 63% BP in NJ, Fig. 4). The genus Mastomys is paraphyletic due to the species Mastomys pernanus that appears quite divergent from the others. The other four species (M. erythroleucus, M. natalensis, M. huberti and M. coucha) form a highly supported clade (100% BP, DI = 10) of closely related species (sequences diverge by 10% at the most), a finding consistent with the MP and NJ analyses based on amino acids. The sister group of this clade is represented by Mastomys verheyeni with a Decay Index of 6, but with very low bootstrap support (57% in MP; 53% in NJ, Figs. 2–4). The relationships within the four closest species were not clearly resolved with the global analyses (polytomy between M. coucha, M. huberti and the grouping M. natalensis/M. erythroleucus), while they were using all the substitutions: the tree obtained was integrated in the global phylogeny. In ML (global model), the relationships are incongruent with those recovered in MP analysis but when using the model for the Mastomys data set, the relationships are the same that in MP analysis, i.e. Mastomys coucha is the first species to diverge, then M. huberti, whereas M. natalensis and M. erythroleucus are sister groups.
3.5 Relative-rate tests
Synonymous (Ks) changes did not show significant differences between the species within the Praomys group. However, Ka comparisons showed that only Mastomys pernanus was quickly evolving (P < 0.05). As the RRTree test is rather sensitive to the rate of species used as outgroups 〚31〛, we also applied the test using Mega 〚32〛. These results obtained confirm the RRTree test when Apodemus or Rattus are used as outgroups; however, when using Mus as outgroup, no taxon show significant difference of evolution rate. Consequently, when applying a molecular clock to estimate the separation dates between the species and genera within the Praomys group, M. pernanus was not taken into account.
4 Discussion
4.1 Genetic distances
The genetic distances observed within the Praomys group are similar to those known between other rodent genera, which were found to span a wide range. For example, between 9 and 20% sequence divergence has been reported among some caviomorph genera 〚35〛, while between 6 and 15% sequence divergence has been found among European muroids 〚34〛.
4.2 Phylogenetic analyses
The results of our phylogenetic analyses are consistent with expectations based on previous studies. Although its dental structure has been reviewed 〚10, 15〛, the affinities of Zelotomys have not been discussed on the basis of biochemical techniques. Jaeger and Davis concluded that it was close to the Praomys group 〚10, 15〛, a proposal confirmed in our study. Similarly, Colomys was proposed for inclusion within the Praomys group in a recent molecular study 〚16〛, while, on the basis of its long feet, and because of its semi-aquatic habits, it has often been compared with Malacomys 〚36, 37〛. Zelotomys and Colomys appear closely related here, as proposed by Misonne on the basis of dental characters 〚4〛, but their sister group was not unambiguously identified, although the species M. verreauxii could represent a good candidate. Heimyscus fumosus was considered a member of the genus Hylomyscus, before Misonne 〚4〛 created a new genus for this species. Our study confirms that Heimyscus is quite divergent from Hylomyscus and that it is probably more closely related to Praomys. Moreover, the genus Myomys was already proposed to be paraphyletic 〚14, 16, 17, 19〛. The close affinity of Myomys albipes with Stenocephalemys was demonstrated by morphometrical, chromosomal, morphological and molecular analyses 〚14, 16, 17〛. Myomys fumatus was found to be closely related to Mastomys 〚38〛 on the basis of chromosomal and protein data. Other Myomys species have been proposed to be related to Praomys (e.g. Myomys daltoni and Myomys derooi), or to Mastomys (Myomys verreauxii) 〚19〛. Were these relationships confirmed, this polyphyly of Myomys should lead to nomenclatural revision of the genus. For instance, Myomys fumatus would join Mastomys 〚38〛, Myomys daltoni and Myomys derooi would have to be placed within Praomys, while Myomys yemeni and Myomys albipes would join Stenocephalemys. The species Myomys yemeni, which lives on the Arabian Peninsula, also clusters within the Praomys group. Its apparently weak divergence from purely African species such as Myomys albipes and Stenocephalemys spp. suggests that M. yemeni only recently established in the Arabian Peninsula. Myomys verreauxii, the only South African species, is the type species of the genus and would retain the name ‘Myomys’.
In our study, the genus Praomys is found paraphyletic. In earlier studies based on DNA/DNA hybridisation and morphometrics, the monophyly of this genus was questioned, based on the high divergence between the P. jacksoni and P. tullbergi groups 〚6, 18〛. The two well-supported groups (P. jacksoni/P. tullbergi) distinguished here, have been identified in other analyses, using chromosomal 〚39, 40〛, palatal ridges 〚41〛, and craniodental characters 〚18, 20, 42〛. Here the P. jacksoni group includes the species P. jacksoni and P. degraaffi, which is congruent with morphometrical and morphological studies 〚20, 43〛. According to morphological characters 〚20〛, the sister taxon to the P. jacksoni group is Myomys daltoni. The P. tullbergi group includes P. tullbergi, P. misonnei, P. hartwigi obscurus and P. sp. nov. 〚21〛, which is also congruent with analyses based on morphological and morphometrical data 〚20, 42〛.
Although Mastomys, as currently recognised 〚1〛, appears to be paraphyletic, four of the Mastomys species (M. erythroleucus, M. natalensis, M. huberti and M. coucha) form a strongly supported clade. These species have often been found to be closely related 〚6–8, 44〛. Based on chromosomal and DNA/DNA hybridisation data, M. coucha was the first species to diverge 〚6, 7〛 while M. natalensis and M. huberti were found to be the most closely related ones 〚7〛. However, using protein electrophoresis data on specimens from Senegal, M. huberti appeared closer to M. erythroleucus than to M. natalensis 〚44〛, but this pattern of relationships may have been obscured by the much reduced genetic variability of M. natalensis at the edge of its distribution area. Our analysis confirms that M. coucha is the sister group of the three other species. However, M. erythroleucus is placed here as the sister taxon of M. natalensis. Mastomys verheyeni seems to be the sister group of these four species. It is easily distinguishable morphologically from the other species 〚5〛, but its phylogenetic position has never been discussed. Finally, the rare Mastomys pernanus was included for the first time in a phylogenetic analysis. Van der Straeten 〚45〛 first mentioned its high morphometrical divergence from Mastomys and questioned its affinities. Our study confirms that this species should no longer be considered a member of Mastomys.
Malacomys verschureni was thought to show affinities with Praomys 〚18, 46〛, and indeed has been often labelled as Praomys within museum collections 〚46〛, but had not been included in any phylogenetic study so far. Here we confirm that this species belongs to the Praomys group, with an especially close relationship to Heimyscus.
The incongruences between the MP, ML and NJ trees and between the nucleotide-based and amino acid-based analyses concerning the basal nodes suggest a telescoping in time of the cladogenetic events, which may have resulted from an adaptive radiation (see below).
4.3 Divergence times
Using the available fossil record, a molecular clock has been calibrated in murid rodents for cytochrome b 〚47〛, based on the divergence date between Mus and Rattus and the transversion rate at third codon position. Palaeontological data suggest that this split occurred around 10–14 Myr 〚48–50〛. There are 70 transversions at the third codon position between Mus musculus and Rattus norvegicus. Taking a mean divergence time of 12 Myr, the rate of third position transversion change is 1.53% per Myr. Using this transversion rate among the taxa under consideration, estimates of divergence times for the cladogenetic events are provided (Table 2). However, a recent molecular study proposed that the Mus–Rattus split may have occurred 23 Myr ago 〚51〛. Thus, divergence dates are also proposed for a split of Mus–Rattus at 23 Myr (Table 2); nevertheless, only the divergence dates based on the split at 12 Myr will be discussed further in order to compare with previous studies based on this dating.
Estimates of divergence times using third position transversion changes numbers for the complete cytochrome b gene dataset, and based on a Mus–Rattus split at 12 and 23 Myr (cf. text), using a rate of third position transversion change of 1.53% per Myr and 0.80% per Myr, respectively. Events are listed as they appear in Fig. 2.
| Number of third position transversion differences (mean) | Divergence time estimates (Myr) according to the Mus–Rattus split at: | ||
| 12 Myr | 23 Myr | ||
| Apodemus / Praomys group | 66.1 | 11.4 | 21.7 |
| 1st divergence within Praomys group | 49.6 | 8.5 | 16.3 |
| 2nd divergence (Ethiopian clade / Praomys, Myomys, Hylomyscus) | 46.9 | 8.1 | 15.4 |
| P. tullbergi group / M. daltoni, P. jacksoni group | 30.5 | 5.2 | 10 |
| Myomys daltoni / Praomys jacksoni group | 28.5 | 4.9 | 9.4 |
| Within Praomys tullbergi group | 12.5 | 2.2 | 4.1 |
| Within Praomys jacksoni group | 20.5 | 3.5 | 6.7 |
| M. yemeni / Ethiopian clade | 44.9 | 7.7 | 14.8 |
| Within Ethiopian clade (M. albipes-Stenocephalemys) | 13.4 | 2.3 | 4.4 |
| M. verreauxii / Colomys + Zelotomys | 38.7 | 6.6 | 12.7 |
| Colomys / Zelotomys | 23 | 3.9 | 7.6 |
| M. fumatus / four Mastomys species group | 42.6 | 7.3 | 14.0 |
| M. verheyeni / four Mastomys species group | 41.2 | 7.1 | 13.5 |
| M. coucha / M. erythroleucus, M. huberti, M. natalensis | 17.3 | 3.0 | 5.7 |
The divergence time separating the Praomys group from Apodemus is estimated here at 11.4 Myr. This date is older than the estimate of 8 Myr for the emergence of the Praomys group from the Mus lineage based on DNA/DNA hybridisation data 〚6〛. The same concerns the estimation of divergence between the genera of the Praomys group (8.5 here vs 4 Myr via DNA/DNA hybridisation data 〚6〛). Among the Praomys group, the resolution is weak above the species level while the node suggesting that Apodemus is the sister taxon of this group is quite well supported, although much older. One explanation would be a deficit of information due to saturation, but the saturated substitutions were removed and the tree length distributions were significantly skewed, indicating a phylogenetic signal in the dataset. Another possible explanation to this apparent telescoping of events is that a phase of extensive diversification did occur in the group, leading to a number of lineages. All the basal splits within the Praomys group are estimated around 7.3 and 8.5 Myr (see Fig. 3). This period of time is rather short and could be an indication that the emergence of the different lineages occurred during an adaptive radiation that took place during Late Miocene. This period was also the one proposed for the split of the Arvicanthini from the rest of Murinae 〚22〛 and for the first appearance of an Antilopinae bovid in Africa as well as migration of endemic Reduncini of genus Kobus in Eurasia 〚52〛. This suggests that some eco-climatic event did favour the radiations and immigrations in Africa. The emergences of Arvicanthini and the Praomys group, calibrated in the same manner, are contemporaneous, and using a molecular clock calibrated on 23 Myr for the Mus–Rattus split would maintain this contemporaneity.
The fossil record is rather scarce in the Praomys group: the earliest records are from Laetoli (Tanzania) dated at 3.7 Myr 〚53〛 for a species of Mastomys, 3.3 Myr for a species of Praomys in Hadar (Ethiopia) 〚54〛 and around 3 Myr for Zelotomys in Gcwihaba (Botswana) 〚55〛. The emergence of the genus Mastomys is estimated here (Table 2) at 7.1 Myr, a date that is compatible with the age of the Laetoli fossil. The estimate of 3 Myr for the emergence of the four Mastomys species (erythroleucus, natalensis, huberti and coucha) is much older than the one of 1 Myr calculated from DNA/DNA hybridisation data 〚6〛. This date of separation may appear remote in comparison with the great morphological similarity between these four Mastomys species 〚8, 56, 57〛, but is more in agreement with their karyotypic differentiation 〚7, 8, 56〛. The divergence date of the two Praomys groups is estimated at 5.2 Myr, while the divergence between the species within each groups is much more recent (i.e. 2.2 Myr and 3.5 Myr for the tullbergi and the jacksoni groups, respectively). The divergence within the Ethiopian clade Stenocephalemys–Myomys albipes is estimated around 2.3 Myr. Within this clade, Fadda et al. 〚16〛, in a molecular analysis based on 16S sequences, found quite similar divergence times (with a time scale calibrated on 12 Myr for the Mus–Rattus split): 1.3 Myr between M. albipes and Stenocephalemys griseicauda against 1.9 Myr in our analysis and 2.24 for the clade versus 2.4 Myr here. Also, Fadda et al. 〚16〛 found between 5.9 and 6.4 Myr for the split of Myomys daltoni from the Ethiopian group, which is less than the 8 Myr value found here.
The date of divergence of this Ethiopian clade (Myomys albipes–Stenocephalemys) is approximately the same as the one estimated for the diversification between Mastomys species or within the tullbergi group, comprising sibling species; however, the species of the Ethiopian clade are so distinguishable from each others that they were not included in the same genus. This is an additional example where rates of morphological evolution do not correlate with rates of molecular evolution. This observation was early highlighted by Wilson and his co-workers 〚58–60〛, and subsequent molecular studies in species groups that have undergone recent adaptive radiations, such as African rift lake cichlids 〚61〛, columbines 〚62〛, and the Hawaiian silversword alliance 〚63〛, have documented marked incongruities in rates of morphological and molecular evolution. A proposed resolution to this paradox has been the conjecture that morphological evolution proceeds via diversification in regulatory loci, and so that phenotypic evolution may correlate better with regulatory gene divergence 〚64〛. Such hypotheses would worth be tested on African rodents in which the patterns and processes of evolution appear highly diversified.
Acknowledgements
We are grateful to the numerous field collectors listed in Table 1, as well as to the generous providers of tissue specimens: F. Catzeflis, D. Kock, S. McLaren, F. Dieterlen, R. Hutterer, A. Hoffman. Our thanks also go to Nicolas Vidal and Jean-François Ducroz for their valuable help and comments, as well as to the two referees who commented on our manuscript. This work was supported by the SSM and the EA 2586 (MNHN, Paris).
Version abrégée
Le groupe Praomys représente un ensemble de genres de rongeurs murinés, essentiellement africains, dont le contenu exact ainsi que les relations phylogénétiques inter et intra-génériques sont jusqu’à présent débattus. Alors que les genres Heimyscus, Hylomyscus, Mastomys, Myomys, Praomys et Stenocephalemys sont des représentants indiscutables de ce groupe, certains auteurs ont proposé qu’y soient également ajoutés Zelotomys, Colomys et l’espèce Malacomys verschureni. Par ailleurs, la monophylie de genres tels que Myomys et Praomys a été plusieurs fois mise en cause, de même que la position de certaines espèces dans ces genres ou dans d’autres du groupe (par exemple, Mastomys pernanus). Afin de tenter de résoudre certaines de ces incertitudes, nous avons réalisé une reconstitution phylogénétique de ce groupe à partir des séquences complètes du gène mitochondrial du cytochrome b d’un échantillon représentatif des espèces de l’ensemble des genres concernés, à savoir : Praomys tullbergi, P. misonnei, P. sp. nov., P. hartwigi obscurus, P. jacksoni et P. degraaffi ; Mastomys natalensis, M. erythroleucus, M. coucha, M. huberti, M. verheyeni et M. pernanus ; Myomys albipes, M. daltoni, M. derooi, M. fumatus, M. verreauxii et M. yemeni ; Hylomyscus stella, H. parvus et H. alleni ; Heimyscus fumosus ; Stenocephalemys albocaudata et S. griseicauda ; Colomys goslingi ; Zelotomys hildegardeae et Malacomys verschureni. Par ailleurs, à partir d’estimations des principales dates de divergence dans ce groupe, calées sur la date supposée de divergence entre Mus et Rattus, nous discutons de la chronologie des événements et des modalités d’émergence de sa diversité, aujourd’hui importante.
Les séquences de 48 spécimens ont été obtenues selon une procédure standard, et traitées avec dix autres issues de Genbank. Une analyse de saturation a été réalisée, montrant que les transitions en première et troisième position de codon étaient saturées. Ces dernières n’ont de ce fait pas été prises en compte dans les analyses. Une analyse de parcimonie (MP) ainsi qu’une analyse de maximum de vraisemblance (ML) et un traitement de la matrice des distances (NJ) ont été effectués sur les séquences nucléotidiques ainsi que sur les séquences d’acides aminés déduites. Le modèle de substitution qui correspond le mieux au jeu de données a été choisi grâce au programme Modeltest ; ce modèle a été utilisé pour les analyses ML et NJ. La solidité des branchements obtenus a été testée grâce à la méthode du ré-échantillonnage et par l’indice de Bremer.
Les séquences obtenues ont montré les caractéristiques classiques du gène du cytochrome b des Mammifères, et une composition en bases nucléotidiques comparable à celle observée chez d’autres rongeurs muroïdés (peu de G – 12,2% – fréquences similaires de A, T et C – 26,7 à 32,1%). Sur l’ensemble du jeu de données, 563 sites ont été trouvés variables (140 en première, 62 en deuxième et 361 en troisième position de codon), dont 423 informatifs pour l’analyse de parcimonie. Les distances génétiques ont varié de 5,3% entre Myomys albipes et Stenocephalemys et de 17,9% entre Mastomys verheyeni et M. pernanus. L’analyse de parcimonie sur les séquences nucléotidiques a abouti à l’obtention de 24 arbres de 1033 pas. Les analyses de maximum de vraisemblance et de distance ont donné un arbre de topologie largement congruente avec celle du cladogramme consensuel. Les analyses basées sur les séquences protéiques ont conduit à des arbres beaucoup moins bien résolus. Les arbres obtenus par MP, ML ou NJ sur les séquences nucléotidiques montrent le groupe Praomys monophylétique lorsqu’il inclut M. verschureni, Colomys et Zelotomys. Parmi les résultats congruents dans les trois types d’analyse, il faut noter la paraphylie de Praomys, qui se trouve séparé en deux groupes d’espèces, le groupe jacksoni (incluant ici P. jacksoni et P. degraaffi) et le groupe tullbergi (incluant ici P. tullbergi, P. misonnei, P. hartwigi obscurus et Praomys sp. nov.). Myomys apparaît quant à lui polyphylétique, avec M. daltoni et M. derooi associés à Praomys, M. albipes à Stenocephalemys, M. fumatus et M. yemeni montrant des affinités moins bien soutenues (le premier avec Mastomys, le second avec le groupe Stenocephalemys–M. albipes). Un clade « éthiopien », réunissant les deux espèces de Stenocephalemys et M. albipes, est très robuste, de même que Zelotomys hildegardeae apparaît comme groupe frère de Colomys goslingi. Le genre Mastomys est paraphylétique, du fait de la divergence marquée de M. pernanus par rapport aux autres espèces du genre considérées ici. Parmi ces dernières, M. coucha, M. huberti, M. erythroleucus et M. natalensis apparaissent s’être différenciées dans cet ordre, M. verheyeni étant le groupe frère de cet ensemble de quatre espèces.
Ces résultats confirment pour certains des données acquises dans d’autres domaines. La monophylie de Praomys et de Myomys avait déjà été mise en cause, et l’ensemble des résultats maintenant accumulés doit aboutir à des révisions nomenclaturales dans ces genres. Ainsi Praomys doit-il, soit inclure M. daltoni et M. derooi, soit être divisé en deux genres (l’un incluant M. daltoni et M. derooi). Par ailleurs, les espèces actuellement rattachées à Myomys doivent être distribuées dans différents autres genres, Myomys verreauxii, l’espèce type du genre, restant éventuellement la seule dans ce genre. Mastomys pernanus doit sans doute être considéré comme appartenant à un autre genre, alors que les cinq autres espèces considérées ici sont assez fortement associées. Toutefois, l’ordre de branchement des différentes espèces demande encore à être confirmé, puisqu’il n’est pas en total accord avec celui déduit d’analyses d’hybridation ADN–ADN, d’électrophorèse des protéines ou chromosomiques déjà réalisées sur ce groupe. La distinction entre Heimyscus et Hylomyscus est clairement confirmée ici, de même que l’inclusion de Zelotomys, Colomys et Malacomys verschureni dans le groupe Praomys. Zelotomys et Colomys apparaissent même relativement étroitement apparentés, alors que la position de M. verschureni est moins claire. Enfin, le seul taxon non africain de l’échantillon, à savoir M. yemeni, originaire de la péninsule Arabique, appartient sans ambiguïté au groupe Praomys, et semble plus précisément être apparenté aux espèces du « clade éthiopien », géographiquement proches. Ceci suggère un passage relativement récent de M. yemeni dans la péninsule Arabique, à partir de l’Afrique de l’Est.
En partant d’une date de divergence entre Mus et Rattus estimée à 12 millions d’années (Ma), et sachant qu’il y a 70 transversions en troisième position de codon entre les séquences du cytochrome b de ces deux taxons, nous avons appliqué un taux de transversion de 1,53% par Ma aux divergences inter-taxons de notre jeu de données. À partir de là, il ressort que le groupe Praomys, tel qu’il est défini dans nos analyses, aurait émergé il y a environ 11,4 Ma. Ensuite, la majorité des groupes (assimilables à des genres) se seraient différenciés entre 8,5 et 7,3 Ma, soit en un laps de temps assez court, à la fin du Miocène. Cette diversification à partir d’ancêtres provenant probablement d’Asie s’apparente par sa rapidité à une radiation adaptative, comme celle qui aurait présidé à l’émergence de la diversité dans la tribu des Arvicanthini, un autre groupe de rongeurs murinés africains. On peut penser que des facteurs éco-climatiques particuliers ont favorisé à cette époque l’entrée de ces immigrants en Afrique, et leur rapide colonisation du continent, accompagnée d’une évolution explosive. À partir de là, en effet, la multiplication du nombre des espèces a été rapide, les espèces actuelles ayant pour la plupart émergé entre 2 et 3 Ma. Il est intéressant de noter que, selon les groupes, cette évolution s’est ou non accompagnée de changements morpho-anatomiques visibles : dans le « clade éthiopien », les différences morphologiques sont telles entre M. albipes et Stenocephalemys spp. que ces taxons ont été érigés en genres différents ; dans le genre Mastomys, comme entre plusieurs espèces du genre Praomys, la différenciation morphologique est restée négligeable, au point que la plupart de ces espèces doivent être considérées comme des espèces jumelles. Des hypothèses quant aux causes de ces différences entre les taux d’évolution moléculaire et morphologique ont été avancées, qui mériteraient d’être testées sur ces rongeurs africains.



