1 Introduction
The shrew genus Crocidura (family Soricidae, order Eulipotyphla [1]) is the most speciose among African mammal genera [2]. These small mammals prey primarily on arthropods, earthworms, and molluscs [3], and are thus ecologically important as secondary consumers [4]. Shrews are relatively abundant, biologically and taxonomically diverse, have a restricted dispersal capacity, and are not subject to hunting pressure. Moreover, because of their short life span, they respond quickly to environmental changes. Thus they have been frequently used as model organisms for phylogeographic studies aimed at understanding factors driving faunal diversification in tropical Africa [5,6] and as bioindicators of habitat change [7–9].
African shrews also bear epidemiological significance because they are increasingly being discovered to be hosts of potentially zoonotic hantaviruses. In Guinea, Crocidura theresae and Crocidura douceti were detected as the reservoirs of Tanganya virus and Bowe virus, respectively [10,11]. In Ivory Coast, Crocidura obscurior was found to carry Azagny virus [12]. Within Tanzania, other shrews such as Myosorex zinki and Myosorex geata have been reported to host the Kilimanjaro virus and Uluguru virus respectively [13].
Identifying Crocidura specimens is notoriously difficult because of the high morphological similarity between species. The ensuing taxonomic uncertainties have resulted in the frequent grouping of taxa in so-called “species complexes” within which the number of species and their geographical distributions are highly debated. This lack of knowledge is a major problem for attempts to delineate appropriate units for biodiversity conservation and for investigations into the distribution ranges of species that may be carriers of zoonotic diseases. Nonetheless, contemporary studies using genetic sequences are beginning to contribute new insight [5,6,14–16].
Very little is known about the taxonomic diversity of shrews and their distribution ranges in Nigeria. In 1983, Hutterer and Happold [17] reviewed the bibliographical data and museum specimens available for this country. Since then, almost no work has been carried out on Nigerian shrews, and specimens from this country are only exceptionally included in recent regional phylogeographic or phylogenetic studies. Up to now, the few published shrew genetic sequences from Nigeria have been generated from a single locality in the southwestern part of the country, Ile Ife [6,18]. In a recent small mammal survey across vast portions of Nigeria, Olayemi et al. [19] listed all their shrew specimens as Crocidura spp. in order to avoid taxonomic misinterpretation.
Nigeria possesses topographical features such as major rivers already shown to be involved in the speciation of other small mammals [20,21]. Therefore, this geographic region not only represents a significant knowledge gap, it also offers a distinct opportunity to contribute to earlier studies on the molecular biogeography of Afrotropical shrews. For instance, the distribution ranges of certain Crocidura clades described in Jacquet et al. [6] that extend only up to the west or only to the east of Nigeria appear to have their boundaries within the country. Furthermore, high altitude could also potentially affect species distribution. That the Jos plateau is home to endemics as the Nigerian Gerbil Taterillus nigeriae, the Ochre Mole-rat Cryptomys ochraceocinereus and the Klipspringer Oreotragus [22] illustrates this point. Another example is the unique montane forest ecosystem on the Mambilla plateau, where a new shrew species Sylvisorex corbeti was described [23] and collection records were made for the first time in Nigeria of other shrews S. camerunensis, S. ollula, and Crocidura attila [24]. Therefore, geographical distribution records, including those for Crocidura shrews, need to be adequately described.
Our objectives in the present investigation are, using mitochondrial gene sequences, to survey the Crocidura taxa present within Nigeria and to describe their distribution according to main rivers and ecological zonation.
2 Materials and methods
2.1 Specimen collection and sequencing
Using Sherman live-traps, the 183 shrew specimens included in this study were collected during 2011–2017 in 19 localities around Nigeria. These localities are distributed across key ecological zones such as the western and eastern flanks of both the Niger and Benue Rivers and along the Guinea savanna and rainforest vegetative belts (Fig. 1). Jos (Jos Plateau, 1280 m above sea level; locality 19) and Ngel Nyaki (Mambilla Plateau, 1650 m above sea level; locality 15) are situated on highlands where several endemic and rare small mammal species have been recorded [22,24]. During the period of the study, each locality was visited at least once in a session of three trap-nights, but certain localities were visited multiply. Traps were set in diverse habitats: inside houses (in kitchens, bedrooms and food storage areas), outdoors in domestic surroundings (including gardens and other sites close to the houses) and outdoors in wild vegetation (e.g., forest, savanna or river bank, depending on the locality). A broad designation of the habitat in which each specimen was captured appears in Appendix A. Our capture methods are described in detail in Olayemi et al. [19], with all voucher specimens preserved in ethanol at the Natural History Museum, Obafemi Awolowo University, Nigeria.
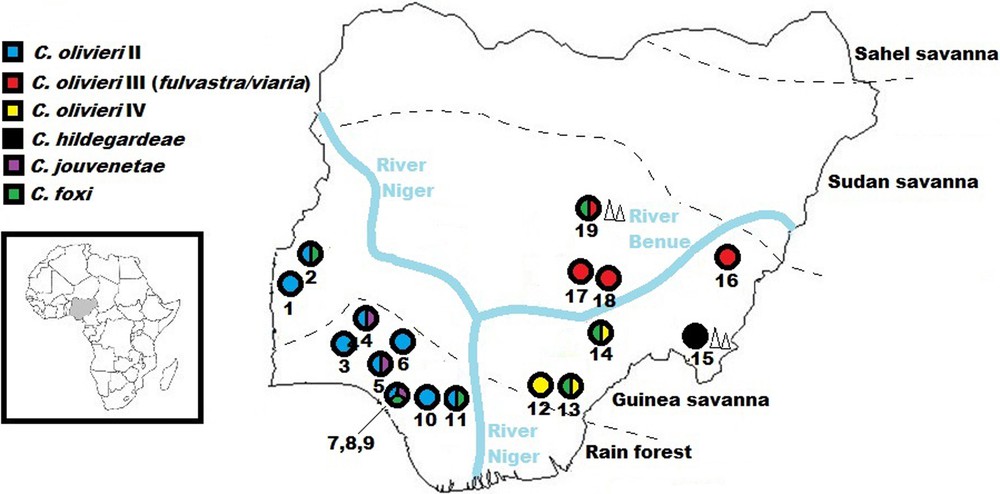
Geographic distribution of Crocidura species and lineages resolved in this study. To the left is a map of Africa, with Nigeria in inset. Colours within the circles indicate the presence of each taxon (but not its relative abundance). Numbers below the circles represent localities as listed: 1) Bwen, 2) Gwanara, 3) Abagboro, 4) Kako, 5) Okeluse, 6) Ifon, 7) Eguare Egoro, 8) Ekpoma, 9) Ebudin, 10) Opoji, 11) Okhuesan, 12) Ndubia, 13) Abakaliki, 14) Onmba Abena, 15) Ngel Nyaki, 16) Mayo Ranewo, 17) Lafia, 18) Obi, 19) Jos. Localities in close proximity (7,8,9) are grouped under the same circle. Geographical coordinates are listed in the Appendix. Triangles designate localities (15 & 19) within highland. Broken lines separate vegetative zones (adapted from Federal Ministry of Environment, Nigeria [25]). It must be noted that patches of derived savanna are scattered over the area classed under rain forest.
No experiments were carried out on living animals. Kidneys were dissected from euthanized specimens and preserved in ethanol. Total DNA was extracted from the kidneys using the Nucleospin Tissue kit (Machery Nagel, Düren, Germany) and the DNeasy blood and tissue kit (QIAGEN, Valencia CA, USA). The cytochrome b gene (Cyt b) was amplified using the following primers: L7 (ACC AAT GAC ATG AAA AAT CAT CGT T) and H15915 (TCT CCA TTT CTG GTT TAC AAG AC) [26]. Initial denaturation was carried out at 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 40 s, and 72 °C for 90 s. Amplicons were Sanger-sequenced and submitted to GenBank with accession numbers from MH996698 to MH996882 (Appendix A).
2.2 Phylogenetic analyses
The obtained sequences were compared to reference sequences in GenBank through a standard Nucleotide BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch). As explained in the introductive section, the taxonomy of many Crocidura species is poorly known. Thus, the aim of our BLAST analysis was simply to be able to attribute each sequence to a given species complex. For each species complex, we then compared our sequences to all relevant published data and/or to unpublished sequences obtained by our team in recent years (details in Appendices B–E). Relationships among sequences were estimated by constructing phylogenetic trees using Neighbour Joining (NJ) phylogenetic analyses. NJ analyses were performed with PAUP 4b10 [27] using the K2P genetic distance. Bootstrap analysis (100 replicates) was used to estimate the robustness of the internal nodes. Note that the aim of this study was not to resolve the phylogeny or taxonomy of the analysed groups, but to provide new information about the distribution ranges of well-differentiated evolutionary lineages using the classical barcoding approach based on mitochondrial sequences in combination with available integrative taxonomy data in the selected groups.
3 Results and discussion
3.1 Notes on individual taxa
3.1.1 C. olivieri species complex Lesson, 1827
The 149 sequences attributed to the C. olivieri species complex based on the BLAST analysis were compared to those published by Jacquet et al. [6]. These authors highlighted the need for taxonomic revision of this complex: the species C. olivieri is polyphyletic and divided into five geographical clades showing allopatric or parapatric distributions, while specimens of C. viaria and C. fulvastra form a single genetic clade. Our Nigerian specimens appear in three different clades: C. olivieri clade II, C. olivieri clade IV and the C. viaria-fulvastra clade III (Figs. 1 and 2). In Nigeria specimens of clade II were only captured west of the Niger River, which seems to represent the easternmost limit of its distribution range. Specimens of this clade were captured both in the Guinea savanna and rainforest zones. We record for the first time the presence of clade IV in Nigeria. This clade was only captured in the Guinea savanna east of the Niger River, which seems to represent the westernmost limits of its geographical distribution. According to Jacquet et al. [6], clade III is widely distributed from Senegal to Cameroon and Chad, but no specimen from Nigeria was previously sequenced. Our specimens fall within the genetic variability already recorded for that species. Clade III specimens were only captured in the Guinea savanna zone. According to Hutterer and Happold [17] C. viaria and C. fulvastra can be differentiated by ventral pelage and tail coloration. Based on these external characters, all our specimens from Nigeria fit more closely with C. fulvastra.
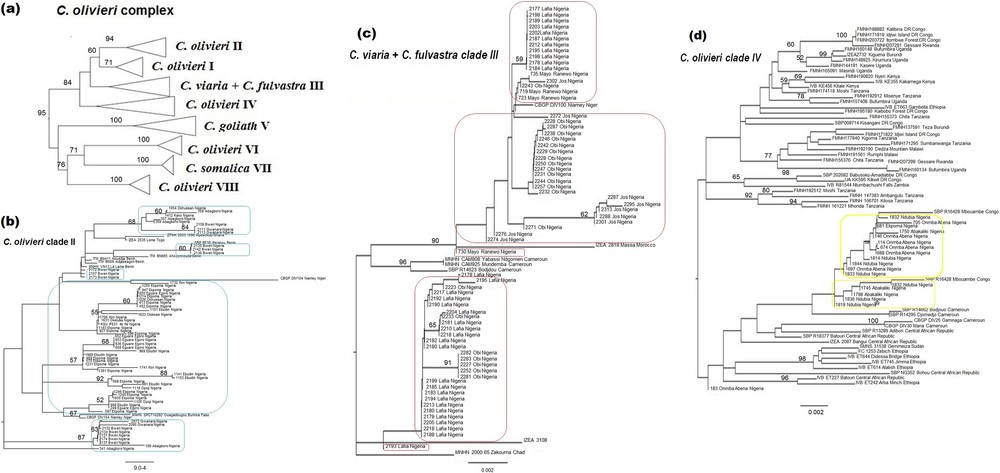
Neighbour joining tree of the C. olivieri species complex based on 1140 bp of the Cyt b gene including the sequences used by Jacquet et al. [6] in their Fig. 2. a: Condensed tree displaying various taxa within the species complex; b: C. olivieri clade II, with Nigerian sequences encased in blue; c: C. viaria + C, fulvastra clade III, with Nigerian sequences encased in red; d: C. olivieri clade IV, with Nigerian sequences encased in yellow.
3.1.2 C. hildegardeae species complex Thomas, 1904
One specimen (NGE777) was attributed to the C. hildegardeae complex based on our BLAST analysis (88–89% identity with C. hildegardeae from Tanzania and Kenya recently published by Stanley et al. [16]). This species complex is in need for revision. Based on 16S mitochondrial sequences, Quérouil et al. [28] and Dubey et al. [29] showed that C. hildegardeae from Burundi, Gabon and Republic of Congo and C. denti from Central African Republic, Cameroun, and Democratic Republic of Congo cluster together and do not form separate genetic clades. In our NJ tree, we included all C. hildegardeae sequences available in GenBank [30] as well as unpublished C. hildegardeae and C. denti from Central Africa, and several representatives of the C. poensis species complex, which is known as the sister clade of the complex C. hildegardeae – C. denti [29]. Our analyses confirm that C. hildegardeae and C. denti are closely related and do not form two distinct monophyletic groups (Fig. 3). Moreover, within C. hildegardeae – C. denti, two main clades can be identified: one grouping specimens from Central Africa (Nigeria, Gabon, Congo Republic and Central African Republic) and one grouping specimens from East Africa (Tanzania, Burundi, Kenya). This analysis confirms the urgent need for an integrative taxonomic revision of C. hildegardeae and C. denti. Neither C. hildegardeae nor C. denti were previously known to occur in Nigeria.
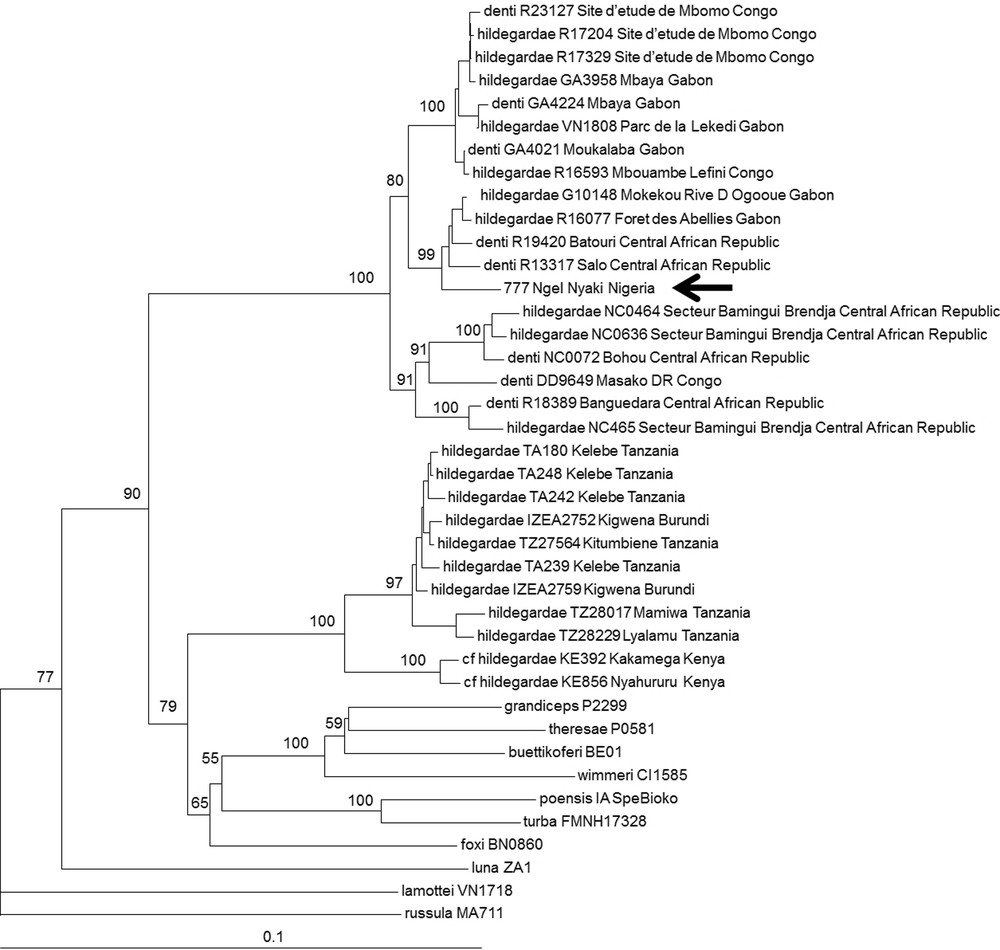
Neighbour joining tree based on Cyt b sequences (1069 bp) of C. denti and C. hildegardeae specimens. Representatives of the C. poensis species complex (C. theresae, C. grandiceps, C. buettikoferi, C. wimmeri, C. turba, C. poensis, C. foxi) were also included in this analysis, and C. luna, C. lamottei and C. russula were used as outgroups. The specimen from Nigeria (NGE777) is indicated by an arrow.
3.1.3 C. cf. jouvenetae Heim de Balsac, 1958
Nine specimens were identified as C. cf. jouvenetae (93% identity with C. cf. jouvenetae published by Vogel et al. [15] based on the BLAST analysis). Crocidura jouvenetae was referred to as a subspecies of C. crossei [31–33]. However, Hutterer [34] considers the larger-sized jouvenetae to represent a separate species. According to Happold [2] C. jouvenetae is found in the rainforest biotic zone of West Africa, in southern Guinea, Liberia, and southern Ivory Coast, and it may also occur in Sierra Leone. Crocidura crossei has a larger distribution: it is known from the rainforest biotic zone, northern rainforest – savanna mosaic and Guinea savanna biotic zone from Guinea and Sierra Leone to western Cameroon. We compared our sequences to all previously published C. crossei and C. jouvenetae sequences [15,29,35] and Kang et al. (unpublished sequences available in Genbank) (Fig. 4). Our results confirm that the complex C. jouvenetae-crossei is in urgent need for integrative taxonomic revision, none of these species being monophyletic. All specimens from Nigeria cluster together with a strong bootstrap support, and they form the sister clade composed of all specimens from Guinea and Ivory Coast. All Nigerian specimens occur in the rainforest zone (Fig. 1).

Neighbour joining tree based on all available Cyt b sequences (402 bp) of C. jouvenetae and C. crossei. The species C. lusitania and C. fuscomurina were used as outgroups. Specimens from Nigeria are encased in purple.
3.1.4 C. foxi Dollman, 1915
Twenty-six specimens were identified as closely related (99% identity) to two specimens from Burkina Faso available in Genbank (DQ305278 and DQ521043) (Fig. 5). These two specimens are identified as “Crocidura theresae” in GenBank and original publications [36,37], but they were later called Crocidura cf. foxi in subsequent studies [15,28]. Crocidura foxi and C. theresae both belong to the C. poensis complex, which was recently reviewed by Nicolas et al. [38]. The comparison of our sequences with those from Nicolas et al. allowed us to unambiguously identify the Nigerian specimens as C. foxi. This species is widespread in Nigeria: we caught it to the west and east of both the Niger and Benue Rivers, and both within the Guinea savanna and the rainforest (Fig. 1).

Neighbour joining tree of the C. poensis species complex (1063 bp of the Cyt b gene), with a special emphasis on the species C. foxi. a: Condensed tree displaying various taxa within the species complex; b: C foxi, with sequences from Nigeria encased in green.
3.2 Biogeographical summary
The number of shrew taxa recorded in this study is small (6) compared to what has been described for Nigeria in the past (24 species in Hutterer and Happold [17], for instance). A reason for our low diversity could be the fact that we used only Sherman traps, which possibly attracts only certain taxa. Surveys in Gabon [39] and in the Congo Democratic Republic [40] demonstrated that pitfall traps are more efficient than Sherman traps for capturing shrews. Also, increasing our effort beyond the three trap-nights we spent in some localities might have yielded more variety. Nevertheless, our survey reports the presence of two taxa detected within Nigeria for the first time: C. jouvenetae and C. hildegardeae.
As could be expected, the Niger River appears to represent a significant physical barrier for several taxa, limiting C. olivieri lineage II and C. jouvenetae on its right bank to western Nigeria (and by extension, West Africa) and C. olivieri lineages III, IV, and C. hildegardeae on its left bank to central and eastern Nigeria (expanding into northern, central and eastern Africa) (Fig. 6). Happold [22] reported 13 mammal species within Nigeria that occurred only on one side of the Niger River, but not on the other. The results of this study also show that the Benue River and/or the drier conditions towards the Sudan savanna in the North might play a combined role with the Niger River in limiting C. olivieri lineage IV and C. hildegardeae to eastern Nigeria.
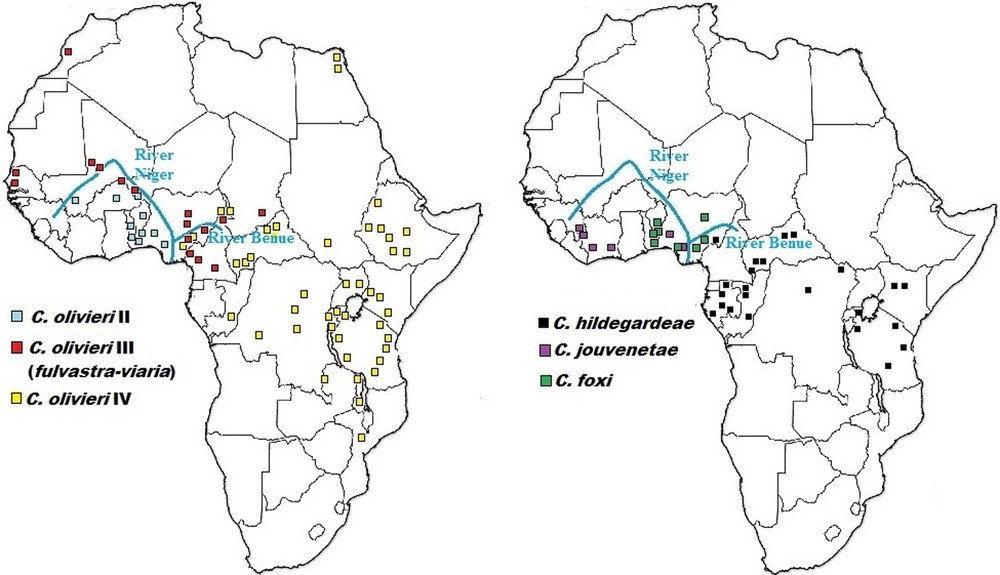
Updated geographical distribution for Crocidura species detected in this study from Nigeria relative to molecular data compiled across Africa by Jacquet et al. [6] (to the left) and Nicolas et al. [38] (to the right).
Elevated topography (represented by the Jos and Mambilla plateaus) apparently does not influence the distribution ranges of the taxa encountered in this study. None of the species was endemic to the Jos plateau. The only C. hildegardeae specimen from Nigeria was found in montane forest within the Mambilla plateau, but specimens from Gabon, Republic of Congo, and Central African Republic were recorded in the lowlands [38].
With regard to habitat, specimens from this study belonging to the C. olivieri lineages and C. foxi were captured indoors, but also outdoors, in a wide range of domestic and wild vegetations (including lowland forest, savanna grassland, banana plants and fallow farmland; see Appendix). Crocidura jouvenetae, on the other hand, was collected outdoors in domestic and wild vegetation (including lowland forest and grassland in forest clearings), while C. hildegardeae was captured only in wild vegetation (montane forest). Many Crocidura taxa have been described as commensal [2]. In Ekpoma, one of the localities sampled in this study, Olayemi et al. [41] detected two Crocidura specimens (later identified by Cyt b sequencing as C. olivieri and C. jouvenetae) with antibodies to the Lassa virus, which causes a deadly viral haemorrhagic fever in humans. They concluded that this was probably the result of a spill-over infection from proximity to the rodent Mastomys natalensis, which is known as the natural reservoir of this virus. These animals are usually found in the same peridomestic habitat. Again, this commensality along with the recent detection of hantaviruses [10–13] and bacteria [42] in Crocidura shrews highlights their zoonotic potential.
Overall our findings, like other recent studies employing molecular data, support the need for taxonomic revision of several taxa within the genus Crocidura. In addition, our survey demonstrates that Nigeria represents the geographical boundary of many Crocidura shrews, and is a region that must not be overlooked in the conservation and management of these animals.
Funding
Fieldwork within Nigeria and part of the laboratory analyses were supported by the European Foundation Initiative for African Research into Neglected Tropical Diseases (EFINTD, grants 1/85/022 & 89 540); and the German Research Foundation (DFG, grant GU883/4-1). Further analyses were funded by the Belgian Science Policy (BELSPO). The funders had no involvement in the design, collection and interpretation of the data.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
We would like to thank Godwin Ehielu and Destiny Aigbomian for their immense assistance in the field. We are also grateful to various persons and institutions that facilitated our trapping around the country including Mallam Isa in Gwanara and Bwen, Drs Shiiwua Manu and Yahkat Barshep of the AP Levenetis Ornithology Research Institute (APLORI), Jos; and Drs Liman Usman and Adamu Ibrahim from the Nasarawa State Ministry of Health.


