1 Introduction
Actin is one of the most abundant and conserved proteins in eukaryotes. The main property of actin is its ability to self-assemble in helical, polar, dynamic filaments, which form one of the main polymers of the cytoskeleton [1]. The actin cytoskeleton reacts to environmental factors like a sensory-motor organ of the cell. Spatially controlled assembly of actin filaments, in response to stimuli, generates cell protrusions called lamellipodia, filopodia and pseudopodia. Motile cells use these extensions to explore the extracellular space and find their way toward their targets, in chemotactic locomotion, embryonic/metastatic cell migration or in morphogenetic processes, like neural cone growth [2,3]. Actin polymerisation is also involved in the movement of endosomes [4], of cortical actin patches in yeast [5], and in the formation of phagosomes [6]. Bacterial pathogens, upon entry in or adhesion to epithelial cells, induce local actin polymerisation [7]. In conclusion, actin assembly is the hallmark of the motile response of cells to extracellular signals.
The extension of lamellipodia is considered as the paradigm of so-called ‘actin-based motile processes’. This thin cellular extension was early recognised as a self-organised system, which continues to display motile behaviour once cleaved off the cell body by microsurgery [8]. Such a process, due to the collective action of a population of filaments growing in a well-organised polarized array, was a complex cell biology problem. Observations of the forward movement of the leading edge of motile cells and of the associated dynamics of actin filaments in the extending lamellipodia clearly pointed out to two striking features. Filaments with their barbed ends pointing toward the leading edge were constantly generated and growing at the plasma membrane during movement, while depolymerising at the rear of the lamellipodium [9,10]. Lamellipodium extension therefore results from the steady-state cycle of actin assembly, the flux of depolymerised actin subunits being used to feed the growth of barbed ends formed at specific sites at the leading edge, in a rapid treadmilling process shown in Fig. 1 [11]. To challenge this view of the thermodynamic basis for actin-based motility, it was necessary to reconstitute actin-based movement from pure components.
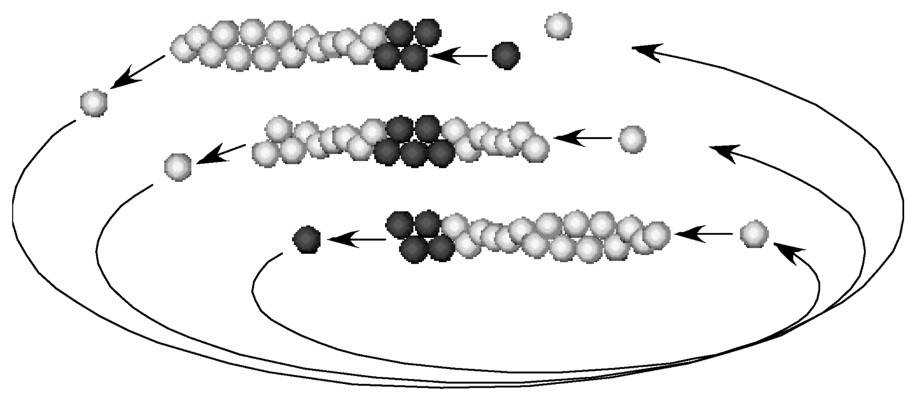
Treadmilling is an intrinsic property of actin filaments in the steady state in the presence of ATP. A pulse of labelled (dark) monomers for a short time makes a mark in the filament. The mark moves toward the pointed end (-end) linearly with time as treadmilling is operating.
Paradoxically, bacterial pathogens have been instrumental in helping to understand a process that takes place in eukaryotic cells. A small number of intracellular pathogens, such as Listeria monocytogenes, Shigella flexneri or Rickettsia (as well as a few viruses like vaccinia virus), induce actin polymerisation at their surface by harnessing the host actin machinery (see [12–15], for reviews). The bacteria continuously generate an actin meshwork or ‘tail’ that promotes their propulsion in the host cytoplasm. A single bacterial factor, the Listeria protein ActA or the unrelated Shigella protein IcsA (also called virG) is required and sufficient for actin assembly and bacterial movement [16–18]. Therefore these pathogens have been rapidly considered as powerful tools to approach the mechanism of actin-based motility. A major progress was accomplished when actin-based motility of Listeria could be monitored in vitro in Xenopus egg extracts [19], then in platelet extracts, brain and Hela cell extracts [20–23]. Depletion and add-back studies of actin regulatory proteins could be performed in the extracts, to identify which ones would be important in motility. Cell extracts were helpful to demonstrate that the Shigella protein IcsA was sufficient to induce actin-based motility of Shigella, and could induce motility in non-pathogenic E. coli [24,25]. Fractionation of platelet extracts led to the identification of Arp2/3 complex as the cellular factor responsible for local stimulation of actin assembly at the surface of Listeria [20], and the important discovery that its biological activity required interaction with the Listeria protein ActA [26]. Recruitment and activation of Arp2/3 complex by Shigella was also demonstrated using cell extracts [27]. Following these achievements, the Arp2/3 complex was recognised to play an important role in lamellipodium extension, as well as in a variety of motile processes. Electron microscopy analysis of the structure of the actin filament array in the lamellipodium showed that the filaments were branched at 70° angle, the barbed ends of the mother and daughter filaments facing the leading edge of the cell, and Arp2/3 complex was localised at the branches [28].
The full reconstitution, from a minimum set of pure components, of actin-based motility of Listeria or Shigella or any particle able to initiate the cascade of events leading to local activation of polarized actin assembly, was a challenging issue. Its achievement, two years ago [29], has opened new perspectives both in the experimental approach of the mechanism of production of force by vectorial assembly of a polymer, and in the potential ways to overcome the virulence of pathogens using actin-based motility for efficient spreading (Fig. 2).

Phase contrast observation of actin-based movement of Listeria and N-WASp derivatised bead in the reconstituted motility medium. The medium contains 8 μM F-actin, 4 μM ADF/cofilin, 2 μM profilin, 50 nM Capping Protein, 90 nM Arp2/3 complex, 0.2 μM VASP. (a) Bacteria (dark) and (b) bead (white), are propelled by ActA-activated stimulation of filament branching and insertional polymerisation of actin, leading to the formation of an actin tail (grey).
2 Rationale for reconstitution of actin-based motility: functions and players in movement
2.1 Actin treadmilling
Two essential biochemical functions appear required to maintain a stationary movement: (1) filaments exposing free barbed ends able to grow actively must be created and maintained at specific sites; (2) growth of these filaments at the specific sites must be made possible by the maintenance of a concentration of monomeric actin well above the critical concentration for barbed end assembly. These two functions are mediated by different proteins: (1) the Arp2/3 complex is responsible for the constant generation of new filaments at a surface; (2) proteins which regulate the treadmilling of actin filaments maintain the flux of subunits i.e. provide the ‘fuel’ feeding this engine.
Genetic identification of proteins that are involved in cell motility and biochemical analysis of proteins that control the dynamics of actin filaments [30,31] have helped to elaborate a minimum reconstituted medium that would be made of pure proteins and would support sustained actin-based movement of Listeria and Shigella [29]. Three essential components in the motility medium and their properties are listed below.
2.2 Actin depolymerising factor
Actin Depolymerizing Factor (ADF)/cofilin is a small (18 kDa), ubiquitous, conserved actin-binding protein, which is responsible for the rapid turnover of actin filaments in motile regions of the cell. ADF/cofilin interacts with both F- and G-actin in the ADP bound form preferentially. Upon binding to F-ADP-actin, ADF/cofilin modifies the structure and the assembly–disassembly rate parameters. ADF-bound filaments become more dynamic due to a large increase in the rate of depolymerisation from the pointed end, which is the rate-limiting step in the treadmilling cycle. A new steady state of actin assembly is established in the presence of ADF, in which a high steady-state concentration of monomeric ATP-G-actin supports fast barbed end growth, to balance ADF-induced fast pointed end depolymerisation (Fig. 3). Hence ADF plays a role in the fast growth of barbed ends in the steady state, which powers actin-based motility [32].

Treadmilling is accelerated by ADF. ADF-decorated filaments display enhanced depolymerisation from the pointed ends (the rate limiting step in the treadmilling cycle). The steady-state concentration of ATP-G-actin is increased, so that fast barbed end growth matches fast pointed end disassembly.
2.3 Capping proteins
Capping proteins block actin assembly–disassembly at the barbed ends. In the steady state, barbed end-capped actin filaments can only depolymerise from their pointed ends. If a large proportion of filaments are capped at their barbed ends, the concentration of monomeric actin settles at a value very close to the critical concentration at the pointed end. The few remaining non-capped barbed ends grow individually at a high rate so as to balance the flux of depolymerisation from all the pointed ends. Treadmilling therefore is funnelled [33]. Capping proteins synergise with ADF to enhance actin-based motility. Any capping protein (CP, the homologue of CapZ in non muscle cells, or CapG, the macrophage capping protein, or gelsolin) is equally efficient in the motility medium [34].
2.4 Accessory proteins: profilin, VASP, alpha-actinin
Profilin interacts with ATP-G-actin specifically. The profilin–actin complex productively associates with barbed ends exclusively, hence profilin cooperates with ADF to enhance the processivity of the treadmilling of actin filaments [35], and improves actin-based motility; however, it is not specifically required. Interestingly, this synergy between ADF and profilin is an example of biological complexity, in which the individual properties of two proteins combine to generate a new behaviour [36].
VASP is a focal adhesion protein, also present at the leading edge of motile cells [37–39], which binds to a central proline rich region of ActA. VASP is almost essential in movement of Listeria [29] and comet tail structural organisation, but does not affect Shigella movement. The molecular mechanism by which VASP enhances motility is currently unknown [40].
Alpha-actinin is not required for bacterial actin-based motility, but its ability to crosslink the filaments is important to anchor the actin tail of the bacteria in the background meshwork that forms the medium. By preventing the drift of the bacterium+tail, alpha-actinin makes the measurements of bacterial speed easier and more accurate.
2.5 Arp2/3 complex
Arp2/3 complex is a conserved seven-subunit complex comprising in particular the Actin related proteins Arp2 and Arp3. The Arp2/3 complex is the downstream target of a variety of signalling pathways leading to actin polymerisation (see [41] for a review). It multiplies actin filaments by branching when it interacts with an activator. The Listeria protein ActA directly activates Arp2/3 complex [26]. The Shigella protein IcsA does not activate Arp2/3 complex, but recruits N-WASp [27,42], the ubiquitous homologue of WASp/Scar family proteins, which are the natural activators of Arp2/3 complex in eukaryotic cells (see [43] for a review). Hence both Listeria and Shigella use Arp2/3 complex to generate new actin filaments at their surface upon activation.
A consensus has not been reached yet concerning the mechanism by which actin polymerisation is stimulated by Arp2/3 complex. There is general agreement however that Arp2/3 complex multiplies the filaments locally by branching either from their sides [44] or from their barbed ends [34,45,46], and is responsible for the dendritic filament array observed in the lamellipodium [28].
3 Spatially controlled activation of Arp2/3 complex: WASp family proteins and ActA
3.1 WASP family proteins
The Shigella protein IcsA is required and sufficient for bacterial motility. IcsA binds N-WASp but not WASp, the homologue of N-WASp in hematopoietic cells [27,42]. WASp family proteins (see [43] for a review) have a multimodular structural organisation. WASp and N-WASp contain in particular a WH1 (WASp homology 1) domain, a basic region that interacts with PIP2, a GBD (G protein binding domain) that specifically interacts with the small G-protein Cdc42 in the GTP bound form, a proline-rich region that binds SH3 domains of Grb2, Nck and some tyrosine kinases, and a conserved C-terminal region (VCA), which binds one G-actin molecule and Arp2/3 complex. The isolated VCA domain constitutively activates Arp2/3-induced actin polymerisation [27,47–49]. In the full-length N-WASp protein, binding of actin and Arp2/3 complex to VCA is prevented due to an autoinhibited fold of N-WASp [50,51]. Binding of PIP2, Cdc42-GTP, Grb2 to their respective targets on N-WASp promotes a structural change of N-WASp, exposing the VCA domain for association with G-actin and Arp2/3 complex, leading to the stimulation of filament branching [48,52–54]. The VCA domain binds one molecule of G-actin in a profilin-like fashion via its V region (verprolin homology), i.e. the VCA-actin complex can productively associate with a barbed end. VCA-actin in addition binds Arp2/3, making a ternary complex, which is the active branching complex. Binding of VCA to Arp2/3 complex enhances binding of ATP to Arp2 specifically [55]. The binding of ATP to Arp2 – not to Arp3 – is required for filament branching by Arp2/3 complex [55]. Filament branching implies the interaction of the ternary G-actin–VCA–Arp2/3 complex with a filament, leading to the initiation of a branch (Fig. 4).

Model for lamellipodium extension. Branching of filaments from their barbed ends is catalysed by WASP family proteins activated at the plasma membrane. Association of WA with G-actin and Arp2/3 complex yields a branching complex, able to interact with a filament barbed end to initiate a branch. Growth of the branches pushes the membrane. Second generation branching can occur if a growing barbed end associates with another branching complex formed at the plasma membrane. This autocatalytic process generates an arborescent filament array.
IcsA mimics Cdc42 in relieving the autoinhibited fold of N-WASp, allowing the VCA domain to catalytically induce filament branching at the surface of Shigella. The interaction between IcsA and N-WASp is so strong that N-WASp remains associated with the bacterial surface during movement of Shigella [27]. Similarly, N-WASp remains associated with the surface of rocketing endosomes during movement [4].
3.2 Listeria protein ActA
The Listeria protein ActA is also required and sufficient for bacterial motility. Two domains of ActA play defined functional roles in movement. The N-terminal domain (residues 1–160) is sufficient to activate Arp2/3 complex [56]. ActA uses the same molecular mechanism as the VCA domain of N-WASp: it binds one G-actin molecule and Arp2/3, the resulting ternary complex stimulates actin polymerisation in vitro by branching the filaments [45].
3.3 Functionalised polystyrene beads: alternative to bacteria
Polymer beads are a valuable alternative to microorganisms. Polystyrene-based (PS) beads are most often used. They allow easy immobilisation of proteins via hydrophobic interactions, under controllable conditions of surface density of immobilised protein, thus providing a greater control of the physical parameters of movement. PS beads of sizes between 50 nm and 10 μm have been successfully used in reconstituted motility assays [15,57–59]; sizes of 0.2–1 μm are most convenient for light microscopy and move fastest. We have found beads from PolySciences Inc. and Bangs Laboratories Inc. to work equally well. Proteins may be immobilised to beads by adsorption or by covalent coupling.
4 Thermodynamic basis for reconstituted actin-based movement: the motility medium
The different players outlined above act together to support the formation of a steady branched filament array. Constant filament branching and growth occurs at the surface of bacteria or beads by local activation of Arp2/3 complex. In a medium containing ATP as an energy source, filaments maintained in a dynamic steady state in the presence of ADF/cofilin, profilin and capping proteins act as a chemostat buffering a high stationary concentration of monomeric actin (ATP-G-actin+profilin-ATP-G-actin), suitable for sustained barbed end growth. The higher the concentration of capping proteins, the shorter the length of filaments in the actin tail [45]. A steady number of filaments is maintained by the balance between the multiplication of filaments by branching and the cooperative depolymerisation of filaments from the pointed ends fibre by fibre, elicited by ADF.
Importantly, no myosin motor is involved in actin-based motility. The concentration dependence of all components shows an optimum for motility, indicating some interplay between them. Indeed an excess of capping proteins, at a given filament branching activity of Arp2/3 complex, can slow down movement, because filaments are arrested more often than they are created. At optimum concentrations of all components, the particle velocity (several μm/min), the length of the actin tails (a few μm), the trajectories, are similar to those observed in vivo or in cell extracts. The formation of a non-polarized ‘cloud’ of F-actin around the particle is followed by breakage of symmetry [60] in which the cloud becomes a polarized actin meshwork, associated with the establishment of a stationary regime of actin-based movement.
5 Perspectives: open issues in the mechanism of actin-based motility
Important issues remain to be solved concerning the relationship between the biochemical reactions involved in motility and the resulting production of force (see [61] for a review).
Theoretical Brownian ratchet models of actin-based motility have been proposed. They first relied on the actin polymerisation-biased diffusion of Listeria, then on the elastic properties of the actin filament [62]. Another model at the mesoscopic scale relies on the elastic properties of the actin meshwork and the shear stress dues to the growth of the gel at the bacterium surface [63,64]. A microscopic model for force production requires further knowledge of the biochemical properties of the proteins that compose the motile machinery assembled at the surface, and the kinetic analysis of the autocatalytic filament-branching process. Analysis at high spatial (2 nm) and temporal (ms) resolution of the trajectory of a bead or bacterium [65] have shown that filaments are in attached (pulling) and detached (growing and pushing) states, leading to the proposal of a ‘tethered ratchet’ model for actin-based movement [66]. Simulations have been carried out of the growth of a branched actin network against an obstacle [67] and of the resulting force-velocity relation, either in the case of simple nucleation, or of autocatalytic branching at the surface of the particle [68]. The predictions made in these two cases can now be tested experimentally. Clearly the field of actin-based motility is now expanding in interest with the complementary approaches of cell biologists, biochemists, physicists and mathematicians.
Acknowledgements
We gratefully acknowledge the ‘Ligue nationale contre le cancer’ (France) for financial support.



