1 Introduction
The amiloride-sensitive Epithelial Sodium Channel (ENaC) is composed of three homologous subunits α,β and γ, each one consisting of two transmembrane domains and two short cytoplasmic tails [1]. Expressed at the apical membrane of numerous epithelia (kidney, lung and colon), it plays a key role in fluid homeostasis, in blood pressure control and in fluid clearance [2–4]. ENaC activity is highly regulated by both hormonal control (aldosterone and vasopressin) and cellular mechanisms (storage, ubiquitination and degradation) [5–9]. Each C-terminus of ENaC subunits contains PXXP and PPXY (PY motif) sequences, which are candidates for interactions with SH3 (Src Homology 3) and WW (38–40-residue domain with 2 conserved tryptophan) domains, respectively [10–13]. The PY motif of both β- and γ-ENaC participates in negative regulation by interacting with WW domains of the E3 ubiquitin-ligase Nedd4 [9] leading to α- and γ-subunits ubiquitination then to ENaC internalisation and targeting for proteolysis [9,14]. Mutations in the PY motif of β- and γ-subunits as identified in Liddle's syndrome (an autosomal dominant form of salt-sensitive hypertension) abolish the interaction with Nedd4 and are associated with an increased ENaC activity [15–18]. However, the role of the PY motif of the α-ENaC subunit is not physiologically established and it is not known whether a mechanism of channel activity regulation, as demonstrated for β- and γ-subunits, can be extended to the α-subunit. The first step to progress in this purpose is to search for ligands of the C-terminus of α-subunit. Using yeast two-hybrid screenings and GST pull-down assays, we demonstrated that α-ENaC interacts with regulating proteins: (i) WWP1, an ubiquitin protein ligase containing WW domains, and (ii) two SUMO/ubiquitin conjugating enzymes, TSG101 and UBC9. Moreover, we also selected Nedd4-2, a new member of Nedd4 family, which has been reported to regulate ENaC activity [19,20]. These biologically coherent data point out the hypothesis of a potential regulation of α-ENaC and target the proteins that must be functionally studied.
2 Materials and methods
2.1 Constructions of two-hybrid plasmids
The human α-PY sequence (QPGPAPSPALTAPPPAYATLGPR) was obtained by annealing of complementary primers containing partial EcoRI and XhoI restriction sites (underlined) AGCCAGGCCCT GCTCCCTCTCCAGCCTTGACAGCCCCTCCCCCT GCCTATGCCACCCTGGGCCCCCGC and GGCGGGGGCCCAGGGTGGCATAGGCAGGGG GAGGGGCTGTCAAGGCTGGAGAGGGAGCAGG GCCTGGCTG, respectively. The rat α-ENaC C-terminus (residues 633 to 716) was amplified using primers CCCGGAGCCGGTACTGGTCTCC and CC CCTCAGAGCGCCGCCA GGCAC. These fragments were cloned in pLEX12 (a modified pBMT116 plasmid, carrying the tetracycline resistance gene). The β- and γ-ENaC C-termini in pBMT116 were generous gifts from O. Staub (‘Institut de pharmacologie et toxicologie’, Lausanne, Suisse). Sequences and expression of hybrid proteins in yeast were verified.
2.2 Yeast two-hybrid system
Two-hybrid screenings were performed as described [21] in L40 strain expressing either LEXA-α-rENaC or LEXA-α-hPY fusion proteins, and transformed with 100 μg of oligo(dT) kidney cDNA library, from rat (established in our laboratory) in pGAD3S2X [21] or from human in pACT2 (Clontech, Ozyme, Saint-Quentin-en-Yvelines, France). Transformation efficiencies were estimated on selective medium lacking tryptophan and leucine (DO-WL). Screening with the α-rENaC bait was performed in addition of 5 mM 3-amino-1,2,4-triazole (3-AT) (Sigma-Aldrich, Saint-Quentin-Fallavier, France). His+ clones were selected after 3–5 days of growth at 30 °C on selective medium lacking tryptophan, leucine and histidine (DO-WLH).
Yeast mating was performed between the L40 haploid strain transformed with pLEX plasmids and its complementary haploid strain AMR70 established with pGAD plasmids. β-galactosidase activity of His+ clones was estimated either on filter, using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), or in liquid assays, using ONPG (O-nitrophenyl β-d-galactopyranoside) on pellet corresponding to 1 ml of culture at OD600 = 0.5. The results are expressed as units as defined by Miller (1972): Unit = (OD420) × (1000/time)×(1/OD600).
Clones were identified by sequencing of PCR-amplified products obtained from yeast plasmids (forward primer CGATGATGAAGATACCCCACC and reverse primers TAATACGACTCACTATAGGGCGA and GAACTTGCGGGGTTTTTCAG for rat and human libraries, respectively). Sequences were submitted to BLAST SEARCH (NCBI).
2.3 In vitro interactions
The α-rENaC C-terminus (residues 633 to 716) was expressed as GST-αrENaC fusion protein after 0.5 mM IPTG induction, from pGEX-2T plasmid (Amersham Biosciences, Saclay, France). After cell lysis by sonication in PBS buffer containing 1% Triton X-100 and 1% anti-protease cocktail (Sigma), GST-fusion proteins were purified on glutathione–sepharose columns (Amersham Biosciences). In vitro interactions were performed for 2 h at 4 °C with 10 μg of recombinant GST-peptides immobilised on Sepharose 4B glutathione beads and either 35S-labelled peptides obtained by transcription and translation in vitro (TNT quicked coupled transcription/translation systems, Promega, Charbonnières, France) in 20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.01–0.2% Nonidet NP-40, 1% anti-protease cocktail or 10 μg of His6-tagged-UBC9 peptide (expressed from pQE80 plasmid; Qiagen, Courtaboeuf; France) in PBS buffer, 0.1% triton X-100. After extensive washings, bound proteins were eluted by GSH and evaluated after SDS-PAGE by either autoradiography or Instant Imager (Packard, Rungis, France) or Western-blots.
2.4 Analysis of the genomic organisation of Nedd4-2
Sequence database searches were performed using BLAST programs at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the ‘Pôle bio-informatique lyonnais’ (http://www.pbil.univ-lyon1.fr) websites. The selected sequences were submitted to SIM4 (pbil) and ModelMaker and EvidenceViewer (ncbi) programs to define putative intron–exon organisation taking into account consensus splice signals.
2.5 Chromosomal location
Chromosomal location of hWWP1 was performed by PCR using the Genebridge 4 Radiation Hybrid DNA panel (UK HGMP Resource Centre). Specific primers were chosen in the 3′ non-coding region of hWWP1: CAGCCAAGAAAAATTGCACAG and CAAATCCAGATCTTTATGAATGAAATG.
3 Results
3.1 The α-rENaC C-terminus interacts with several partners involved in ubiquitin/SUMO pathway
Two yeast two-hybrid screenings were performed on cDNA library either from rat kidney using the C-terminus of α-rENaC (including the PXXP and PY motifs), or from human kidney with a short bait containing the α-PY motif. Among the selected His+ LacZ+ clones, four partners have been found to interact specifically with α-ENaC (no interaction observed with empty pLEX plasmid) as analysed by yeast mating. Two of them are E3 ubiquitin ligases containing WW domains: Nedd4-2 (n=4), for which an interaction with ENaC has been functionally established [19,20] and WWP1 (n=63), for which an interaction with ENaC has been described, but without functional studies [22]. The two other potential partners UBC9 (n=10) and TSG101 (n=2), members of the E2 ubiquitin/SUMO-conjugating enzyme family, are new ENaC-interacting proteins.
3.2 Structural analysis of the α-ENaC-interacting proteins
Nedd4-2, member of the Nedd4 family, consists of an alternative C2 domain (Ca2+-dependent lipid binding domain) implicated in targeting to the membrane, 3–4 WW domains and a HECT (homologous to the E6-associated protein carboxyl terminus) domain responsible for the ubiquitin-ligase activity [19,20]. Compared to Nedd4-2a, also known as KIAA0439 (accession number AB007899) and Nedd4La (AF210730) isoforms, the Nedd4-2 clones selected by our screening correspond to a new isoform of Nedd4-2 (accession number AY312514), which we named Nedd4-2c (Fig. 1). This isoform is characterised by the presence of a new sequence at the 5′ end. This sequence contains an ATG in frame with the previously described start codon in Nedd4La, producing a protein with 120 additional residues and a complete C2 domain (SMART and PFAM programs). Nedd4-2a has a different 5′ sequence, leading to a partial different C2 domain (Fig. 2A). Our Nedd4-2c clones also contain a 60-nucleotide insert located between the first and second WW domains. Nedd4-2 isoforms are encoded by a gene located on 18q21.31. BLAST analysis of Nedd4-2c cDNA sequence against genomic sequence (contig NT_011085.9), followed by the search for putative exons (see materials and methods) revealed that Nedd4-2 gene is made of at least 34 exons spanning around 354 Kbp. The novel 5′ end sequence found in Nedd4-2c and coding for the beginning of the C2 domain corresponds to exons 1 and 2 followed by exons 5, 6, 7 and 9. The 5′ end described in Nedd4-2a corresponds to exon 3, 5, 6, 7 and 9. Nedd4La mRNA contains exons 4, 5, 6, 7 and 9, but the first ATG is in exon 10, leading to an isoform lacking C2 domain. The putative exon 8, evidenced by the web programs, was not recovered in any of the Nedd4-2 isoforms. The 60-nucleotide insert present in Nedd4-2c is encoded by exon 16. Thus, the clones isolated in the two-hybrid screening correspond to the largest isoform encoded by the Nedd4-2 gene, with a complete C2 domain, four WW domains, a 20-residue insert located between WW1 and WW2 domains, and a HECT domain.
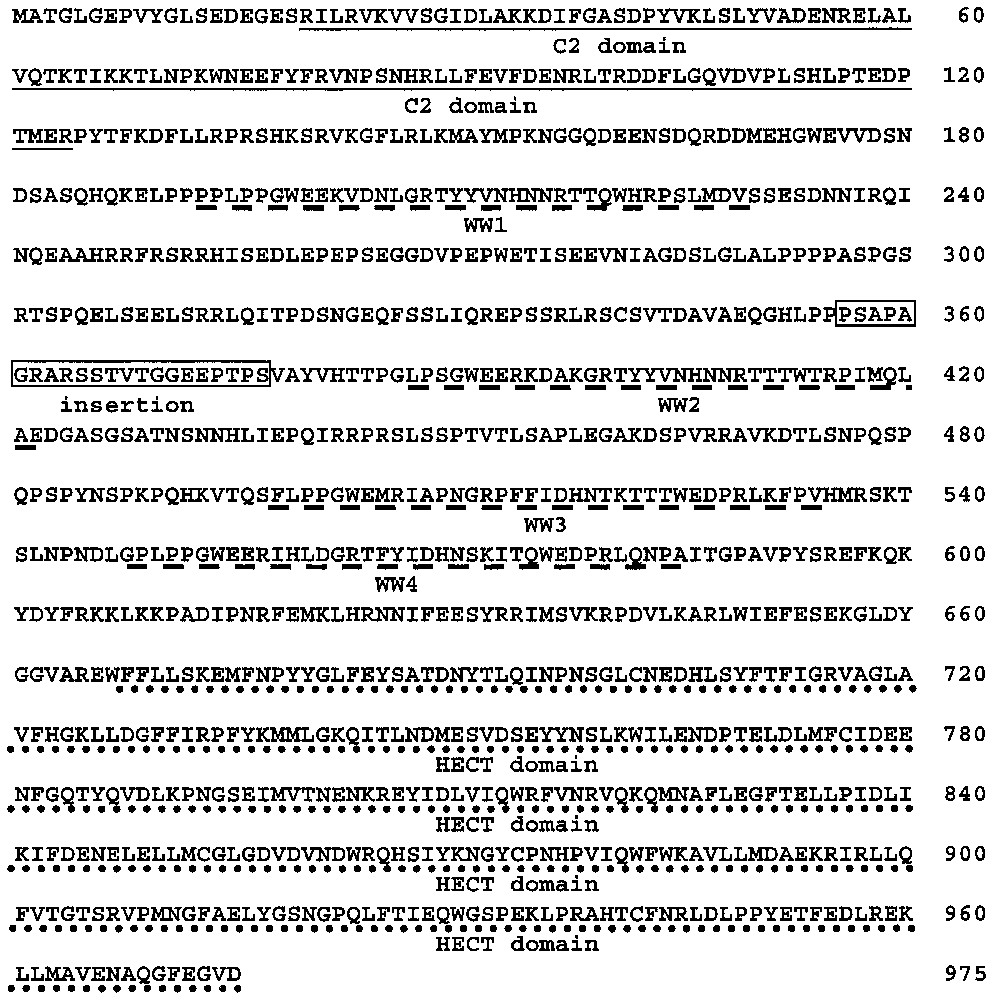
Amino acid sequence of the hNedd4-2c isoform (accession number AY312514). hNedd4-2c is composed of a C2 domain (underlined), four WW domains (dashed line) and a HECT domain (dotted line). This isoform also contains a 16-residues insertion (grey box) located between the first and the second WW domain.
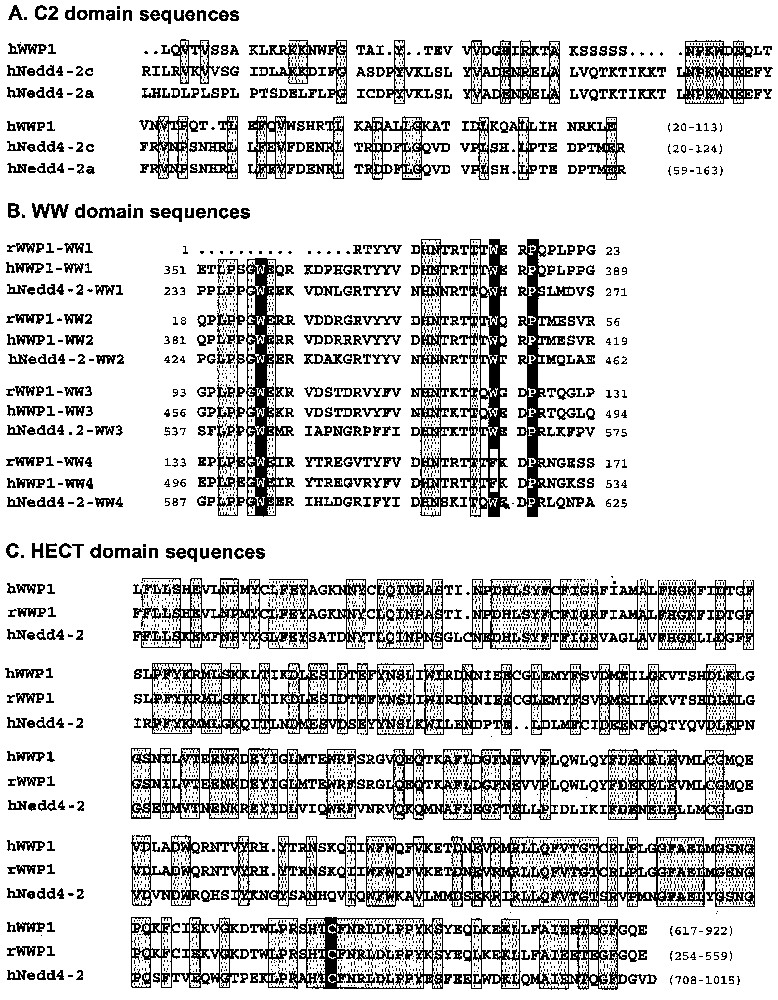
Alignment of functional domains from rat and human WWP1 and human Nedd4-2c (C2, WW and HECT domains). Identical amino acids were included in grey boxes; critical residues responsible for function are in black boxes. Nedd4-2a and Nedd4-2c isoforms differ by the C2 domain.
The WWP1 protein shares structural organisation with Nedd4 family members [22,23]. The WWP1 clones selected by our screenings begin in the middle of the C2 domain and contain four WW domains and a complete HECT domain. This HECT domain contains the cysteine required for the ubiquitin-ligase activity, suggesting a potential enzymatic activity (Fig. 2C, black box). The first three WW domains from rat and human WWP1 have the ‘WWP’ consensus pattern of WW domain (two tryptophan separated by twenty residues and followed by a proline) [24], whereas the fourth WW domain of both rat and human WWP1 lacks the second conserved W (Fig. 2B, black boxes). The human WWP1 gene has been previously located in 3q2.1. However this sequence (AC016962) differed slightly from our WWP1 clones (95% identity), as well as from the one reported by Pirozzi [22]. Moreover, the presence of three stop codons and the absence of intronic sequences suggested that this sequence in 3q2.1 could correspond to a pseudogene. The data we obtained by analysis of Hybrid Radiation DNA Panel allowed to precise a unique location of the human WWP1 gene in 8q21. This location has been recently confirmed by FISH [25].
The clones corresponding to UB2I/UBC9 (NM_ 003345) contain the complete catalytic domain UBCc, usually found in E2 ubiquitin-conjugating enzyme. However UBC9 conjugates an ubiquitin homologue known as SUMO instead of ubiquitin [26]. Like UBC9, TSG101 (U52945) contains an UBCc domain. However, the UBCc domain of TSG101 does not contain the cysteine residue required for the conjugating activity. The TSG101 clones contain almost full-length sequence, beginning six residues after the initiating methionine.
3.3 Beside the PY motif, the C-terminus of α-ENaC contains additional reactive domains
The involvement of the PY motif in ENaC binding activity has been evaluated by the measurement of β-galactosidase activities in the two-hybrid system. Concerning Nedd4-2c, WWP1 and UBC9, the β-galactosidase activities were two to ten times higher in the presence of the complete C-terminus of α-ENaC than in the presence of the PY motif (Fig. 3A). These data confirm the involvement of the PY sequence in the interactions, but show that additional sequences reinforced these interactions, particularly for UBC9. In contrast, TSG101, which was only selected by the C-terminus of α-ENaC, did not interact with PY sequence, suggesting that either the PY motif is not the binding site for TSG101 or flanking sequences of PY are required for the interaction with TSG101.
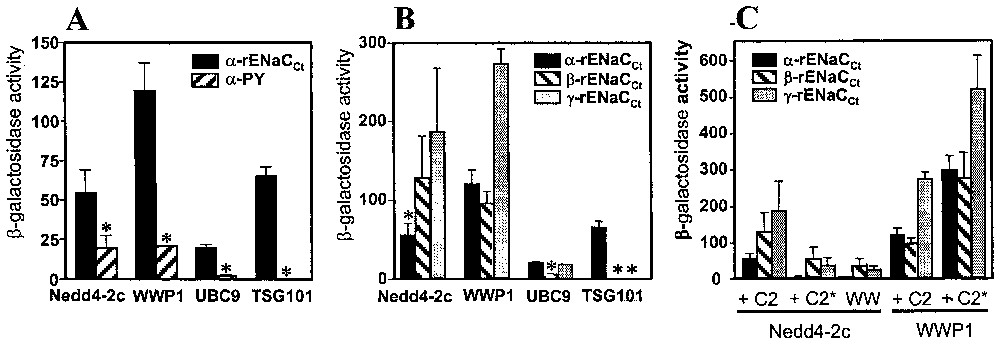
Interaction of the ENaC-interacting proteins with either α-PY motif or α-, β- and γ-ENaC, in the two-hybrid system. Diploid yeast cells were obtained by mating between L40 yeast strain, transformed by either pLEX-C-termini from α-, β- and γ-rENaC or pLEX-α-PY domain and its complementary AMR70 yeast strain, transformed by either pGAD-WWP1, pGAD-Nedd4-2c (containing a complete or a truncated (C2∗) C2 domain), pGAD-UBC9 or pGAD-TSG101. β-galactosidase activities were analysed in liquid assays as described in materials and methods. Results are the mean and standard error of four measurements. Panel A: involvement of the PY motif in the interaction between α-ENaC and the ENaC-interacting proteins; panel B: interaction of the ENaC-interacting proteins with α-, β- and γ-ENaC; panel C: involvement of the C2 domains of WWP1 and Nedd4-2c proteins in the interaction with α-, β- and γ-ENaC.
3.4 Each selected partner displays different interaction affinities with α-, β- and γ-ENaC
As the three ENaC subunits are homologous, we tested the interaction between each ENaC subunit and the four α-ENaC-interacting proteins in the two-hybrid system (Fig. 3B). The highest β-galactosidase activity occurred in the interaction between WWP1 and γ-ENaC, whereas half of this activity value was found in the interaction with α- and β-subunits. The Nedd4-2c isoform interacted better with β- and γ-subunits than with α-subunit, whereas UBC9 interacted mainly with α- and γ-ENaC. Interestingly, TSG101 interacted specifically with α-ENaC, as no binding activity could be detected with β- and γ-ENaC. These results show that the selected proteins exhibit different interaction affinities according to the three ENaC subunits.
3.5 The C2 domains from Nedd4-2c and WWP1 modify the interaction with ENaC
All the Nedd4-2c and WWP1 selected clones contain at least the last two WW domains (WW3 and WW4) with a partial sequence of WW2, suggesting that these domains are required in the interactions. We studied if additional sequences, in particular the C2 domain, might participate in the interactions, since isoforms lacking the C2 domain have been described [20,25]. We tested the effect of the complete C2 domain on the interaction between ENaC subunits and the Nedd4-2c isoform or WWP1 (Fig. 3C). The C2 domain has been found to reinforce the interaction between Nedd4-2c and each individual ENaC subunits when compared to Nedd4-2c lacking the C2 domain. In contrast, the presence of the C2 domain decreased the interaction between the WWP1 protein and ENaC subunits. These results suggest that the presence of the C2 domain could modulate the affinity of the Nedd4 family members for their target proteins.
3.6 In vitro interactions by GST-pull-down assays
Interactions between α-rENaC and the potential partners were analysed in vitro by GST-pull-down assays (Fig. 4). Rat and human WWP1 and rat TSG101 were produced as 35S-methionine-labelled peptides by transcription and translation in vitro and human UBC9 was produced as His6-tagged recombinant peptide. The C-terminus of α-rENaC, expressed as GST fusion protein, retained specifically partial rat (60 kDa) and complete human (115 kDa) WWP1, as well as rat TSG101 (43 kDa) in a higher extend compared to GST (Fig. 4A). Coomassie blue staining and Western-blot revealed that UBC9 (18 kDa) specifically bound to GST-α-rENaC (Fig. 4B and C).
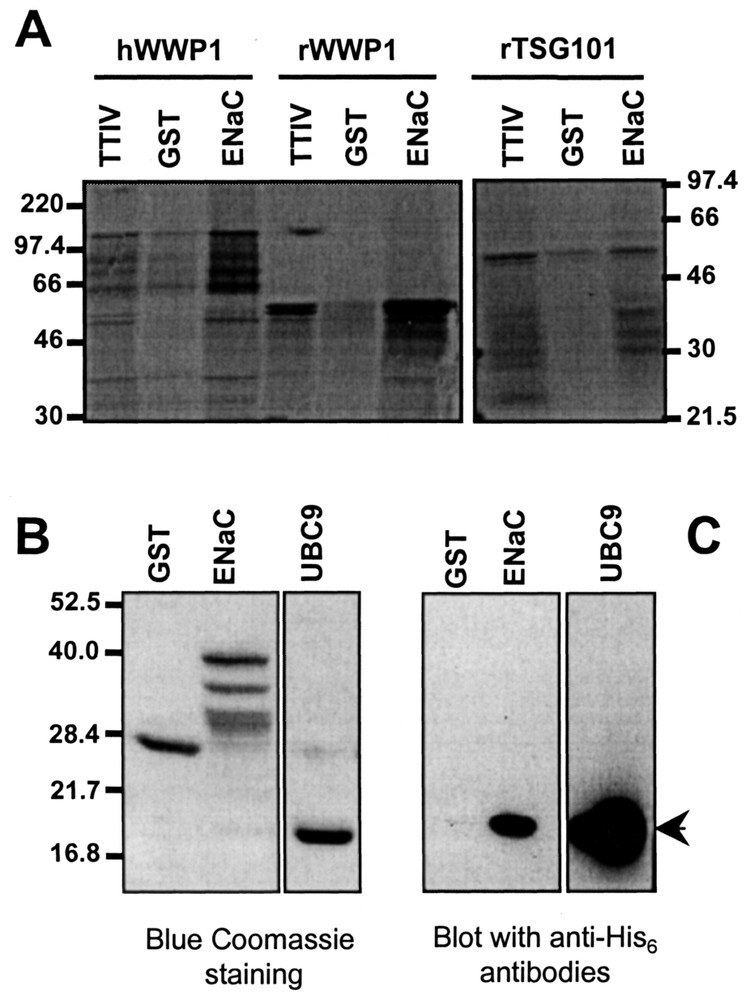
In vitro interaction between α-ENaC and ENaC-interacting proteins by GST-pull down assays. GST and GST-α-ENaC C-terminus peptides were immobilised on sepharose 4B beads and incubated with either rat and human WWP1 and rTSG101 expressed as 35S-methionine labelled peptides or human UBC9 expressed as His6-tagged peptides. After extensive washings, retained proteins were eluted by 10 mM glutathione then analysed by SDS-PAGE. 35S-methionine-labelled peptides were analysed by autoradiography (panel A) and His6-tagged peptide was analysed by coomassie blue staining (panel B) or western-blot using anti-His6 antibodies (panel C).
4 Discussion
Physiological regulation of ENaC relies mostly on trafficking and cell surface stability. However, these processes remain poorly defined and are difficult to study, because of the low abundance of this channel in native tissues. Using the yeast two-hybrid sensitive method to identify α-ENaC-interacting proteins, we selected a new isoform of Nedd4-2 and identified three new potential ligands: WWP1, TSG101 and UBC9. These proteins, which were confirmed to interact with ENaC in GST pull-down assays, could be implicated in trafficking processes mediated by ubiquitin or ubiquitin-like protein (SUMO).
Nedd4-2 belongs to the E3 ubiquitin ligases family, whose members target the ubiquitination process by interacting with specific substrates. The organisation of the human Nedd4-2 gene deduced from the genomic database allowed us to account for isoforms resulting from alternative splicing. Another study contributed to the knowledge of the organisation of the human Nedd4-2 gene [27]. Compared to their proposed organisation, we identified an additional exon 1 containing an ATG in frame with the previously described ATG in Nedd4La. We also identified many additional alternative exons located in the 5′ end. However, starting from exon 9 to the end of the sequence, our proposed organisation is similar to that described by Dunn and collaborators [27]. We isolated a new isoform of human Nedd4-2 (Nedd4-2c, accession number AY312514) characterised by a complete C2 domain, 4 WW domains, a HECT domain and a 20-residue insertion located between the first and second WW domains. The C2 domain of this isoform is encoded by exons 2, 5, 6, 7 and 9 (described as exons 2–6 by Dunn et al. [27]). In agreement with the Nedd4-2a studies reported by Kamynina et al. [19,20], we showed that the Nedd4-2c isoform interacts with the three ENaC subunits but preferentially with β- and γ-ENaC, involving probably the PY motif of ENaC and the WW domains of Nedd4-2c. Moreover, our data revealed that the presence of the complete C2 domain in Nedd4-2c reinforced the interaction with each ENaC subunit. These data suggest the involvement of the C2 domain not only in the recruitment of these proteins to the plasma membrane, but also in the interaction between the enzyme and its substrate. As Nedd4-2 is responsible for the down-regulation of ENaC, the expression of Nedd4-2 isoforms, differing by a C2 domain, a 20-amino-acid insert and a variable number of WW domains, could differently modulate ENaC activity.
The WWP1 protein, another member of the Nedd4 family, has been selected by both α-ENaC C-terminus and α-PY motif in human and rat libraries. Such interaction was previously found by Pirozzi [22] using the COLT method (cloning of ligand target). In agreement with Flasza et al. [25], we found, using a different strategy, that the WWP1 gene is located in 8q21. Although an enzymatic activity is not yet demonstrated, both human and rat WWP1 proteins showed the presence of the highly conserved cysteine required for the ubiquitin-ligase activity in the HECT domain. Several WWP1 mRNAs have been described, resulting from alternative splicing of exons coding for the C2 domain [25]. According to the classification proposed by Flasza et al. [25], our selected WWP1 clones correspond to isoform hWWP1-A. In contrast to Nedd4-2c, the presence of the C2 domain in WWP1 decreases its interaction with ENaC, suggesting that this C2 domain could also modulate the interaction. Consequently, the different WWP1 isoforms differing by the C2 domain could have different substrate affinities. The physiological role of WWP1 in the cell is not well known, although recent studies have reported that WWP1 could interact with transcription factors such as the hematopoietic transcription factors NF-E2 [28] and the lung Krüppel-like Factor 2 [LKLF] [29] and regulate their transcriptional activity.
The second set of selected clones consists on two new ENaC-interacting proteins: TSG101 and UBC9, two proteins related to E2 ubiquitin-conjugating enzymes. Our data showed that TSG101 does not interact with the PY motif but requires another interacting domain specific to α-subunit, as TSG101 does not interact with β- and γ-subunits. Although TSG101 is structurally related to E2 ubiquitin conjugating enzymes, its UBCc domain lacks the critical catalytic cysteine residue essential for conjugation. Originally identified as a tumour susceptibility gene, TSG101 has been recently found to be the mammalian homologue of the yeast Vps23p (vacuole protein sorting) [30]. Both proteins play a key role in regulation of intracellular trafficking, especially in recognition and transport of ubiquitined proteins from the endosome to the vacuole/lysosome. Cells expressing TSG101 mutants exhibit prolonged activation of the EGF receptor-signalling pathway [30]. In contrast to TSG101, UBC9 presents a weak interaction with the PY sequence but this interaction is stronger with the α-ENaC C-terminus, suggesting that the binding site covers a region larger than the PY sequence. UBC9, composed of only one UBCc domain, is required for conjugation of an ubiquitin-related SUMO (Small Ubiquitin-related MOdifier) protein instead of ubiquitin [26]. In contrast to ubiquitination, SUMOylation does not seem to mark proteins for degradation but could function as a antagonist of ubiquitin [26]. UBC9 is expressed in many tissues and is detected in cytoplasm and nucleoplasm, where it plays critical roles in DNA repair, cell cycle regulation and p53-dependent processes [26]. Whereas most SUMO targets seem to be involved in nuclear functions, the GLUT1 and GLUT4 glucose transporters are linked to the SUMO pathway [31].
The identification of these four α-ENaC-interacting proteins, by two-hybrid screenings and interaction studies, provides new insights in the regulation of ENaC activity and targets the studies needed for functional demonstration.
Acknowledgements
We thank O. Staub for generous gift of the β- and γ-ENaC subunits. We are grateful to B. Grandchamp, N. Farman and N. Lemaire for helpful discussions. L. Malbert-Colas is a recipient of a grant from the ‘Ligue contre le cancer’ (France).


