Version française abrégée
Les dernières années ont vu le développement de l'étude moléculaire de l'éléphant africain, jusque là considéré comme une seule espèce : Loxodonta africana (Blumenbach 1797) [1]. Depuis la publication de la première séquence de cytochrome b d'un éléphant de la forme de forêt [2], les phylogénies moléculaires ont relancé le débat de son statut systématique. Des analyses moléculaires [3] ont relancé l'hypothèse selon laquelle deux espèces d'éléphants coexistent en Afrique : Loxodonta africana, l'éléphant de savane et Loxodonta cyclotis (Matschie 1900), l'éléphant de forêt. Cette position était notamment défendue au début du siècle dernier, à une période où la systématique de l'éléphant africain était modifiée en permanence par la multiplication des taxons décrits à partir de variations morphologiques discutables [4–7].
De cette époque, trois taxons ont été retenus, mais jusqu'à aujourd'hui, seuls deux d'entre eux ont été le sujet d'analyses moléculaires : les formes cyclotis et africana ont été étudiées à partir de séquences d'ADN mitochondrial [8] et nucléaire [3], mais rien n'a été fait à propos de « l'éléphant pygmée » de la forêt pluviale africaine : Loxodonta pumilio (Noack 1906). Ce nom n'est pas valide, parce que le type de L. pumilio est un juvénile qui s'est révélé au cours de sa croissance un L. a. cyclotis. De même, l'autre binôme attribué aux petits « éléphants d'eau » décrits par Schouteden [9] : Loxodonta fransseni, mis en synonymie avec L. pumilio, est lui aussi invalidé par l'observation que le type de L. fransseni est un jeune mâle – d'âge D4/M1 – loin d'avoir achevé sa croissance. Aussi bien L. pumilio que L. fransseni sont des nomina nuda, et pumilio est utilisé par simple convenance, d'autant que le taxon n'est pas reconnu dans cet article.
L'existence d'un éléphant de taille naine en Afrique a été défendue de façon récurrente au cours du siècle dernier, bien que notre connaissance à son sujet repose avant tout sur des spécimens de captivité [10,11] ou de collection. Des auteurs l'ont décrit comme une forme de très petite taille de l'éléphant de forêt (hauteur à l'épaule inférieure à 2 m) [12–15], vivant de la Guinée équatoriale à la république démocratique du Congo [16], et dont les mœurs aquatiques ont été postulées [9,17]. D'autres ont nié l'existence de cet animal [18,19], considérant qu'il s'agit plutôt, soit d'individus atteints de pathologies liées à la croissance [20], soit de spécimens juvéniles [20,21] (bien que l'existence d'adultes de taille naine ait été démontrée par ailleurs [22]). Du fait de cette incertitude, certains ont choisi, comme compromis, de faire des éléphants pygmées une sous-espèce : Loxodonta africana pumilio [17,23]. De façon remarquable, les éléphants pygmées semblent afficher le même niveau de différenciation morphologique (discrimination par la taille adulte) et écologique (préférendums écologiques distincts) envers les cyclotis que les cyclotis envers les africana.
Alors que la séparation des cyclotis et des africana en deux espèces semble se dessiner [3,24–26] (mais voir Debruyne et al. (en préparation) pour un autre avis), nous testons ici la validité de la forme pumilio comme troisième espèce africaine. Le débat sur l'existence d'une forme d'éléphants pygmée ayant été entretenu principalement à partir de spécimens de collection de taille extrêmement réduite, il était intéressant de croiser ces hypothèses morphologiques avec des données moléculaires fondées strictement sur le même échantillon. Nous avons donc séquencé un fragment de 1961 pb de l'ADN mitochondrial (recouvrant tout le gène du cytochrome b, l'ARNt Thr, l'ARNt Pro et la région hypervariable 1 de la région de contrôle) pour neuf éléphants de collection enregistrés en tant que « pygmées » et dont les caractéristiques morphologiques, préalablement étudiées [13,22], ont été contrôlées. À ces éléphants nains, huit éléphants de forêt « typiques » ont été ajoutés. Ces échantillons proviennent en majorité du bassin du Congo, dont les populations sont restées inédites sur le plan moléculaire. Enfin, trois éléphants d'Afrique de savane, auxquels s'ajoutent deux éléphants d'Asie (utilisés comme extra-groupes) complètent l'échantillon (Table 1).
Description of the samples from collection/extant specimens
| Taxon1 | Collection | Provider | Geographic origin | Sample | Museum | Morpholo- | In-text reference | Access No. |
| label | type | reference | gical | (Genbank) | ||||
| analysis | ||||||||
| L. africana | africana | Basel Zoo (Switzerland) | South Africa | Hair | – | – | Laa South Africa 1 | AF132528∗ |
| L. africana | africana | Thoiry Reserve (France) | South Africa | Hair | – | – | Laa South Africa 2 | AF132529∗ |
| L. africana | africana | Thoiry Reserve (France) | Namibia | Hair | – | – | Laa Namibia | AF132527∗ |
| L. africana | cyclotis | MNHN (France) | Sierra-Leone | Hair | – | – | Lac Sierra-Leone | AF132530∗ |
| L. africana | cyclotis | Lope Reserve (Gabon) | Lope Reserve, Gabon | Muscle | – | – | Lac Gabon 1 | AY359278 |
| L. africana | cyclotis | Fribourg Museum (Switz.) | Southern CAR | Bone | – | – | Lac CAR | AY359272 |
| L. africana | cyclotis | MNHN (France) | Congo Brazzaville | Bone | No. 1906–450 | [22] | Lac Congo 2 | AY359268 |
| L. africana | cyclotis | MNHN (France) | Cameroon | Bone | No. 1928–122 | – | Lac Cameroon 2 | AY359269 |
| L. africana | cyclotis | MRAC (Belgium) | Bosobolo, DRC | Bone | No. 20333 | [22] | Lac DRC 9 | AY359279 |
| L. africana | cyclotis | MRAC (Belgium) | Uele, DRC | Bone | No. 8203 | – | Lac DRC 3 | AY359271 |
| L. africana | cyclotis | MRAC (Belgium) | Na Bodio, DRC | Bone | No. 8188 | – | Lac DRC 2 | AY359270 |
| L. africana | pumilio | MNHN (France) | Aloombe Coast, Gabon | Bone | No. 1950–728 | [13] | Lap Gabon 2 | AY359265 |
| L. africana | pumilio | MNHN (France) | Congo Brazzaville | Bone | No. 1956–192 | [22] | Lap Congo 1 | AY359266 |
| L. africana | pumilio | MNHN (France) | Yambong, Cameroon | Bone | No. 1956–194 | [22] | Lap Cameroon 1 | AY359267 |
| L. africana | pumilio | MRAC (Belgium) | Bosobolo, DRC | Bone | No. 20330 | [22] | Lap DRC 6 | AY359273 |
| L. africana | pumilio | MRAC (Belgium) | Bosobolo, DRC | Bone | No. 20331 | [22] | Lap DRC 7 | AF517566∗ |
| L. africana | pumilio | MRAC (Belgium) | Bosobolo, DRC | Bone | No. 20332 | [22] | Lap DRC 8 | AY359274 |
| L. africana | pumilio | MRAC (Belgium) | Moma, DRC | Bone | No. 9524 | [22] | Lap DRC 5 | AY359276 |
| L. africana | pumilio | MRAC (Belgium) | Moma, DRC | Bone | No. 7692 | [22] | Lap DRC 4 | AY359275 |
| L. africana | fransseni | MRAC (Belgium) | Mai-Ndombe Lake, | Tooth | No. 3396 | [22] | Lap DRC 1 | AY359277 |
| DRC | root | |||||||
| E. maximus | indicus | MNHN (France) | India | Hair | – | – | Emi India | AF132520∗ |
| E. maximus | indicus | La Palmyre Zoo (France) | Burma | Hair | – | – | Emi Burma | AF132521∗ |
1 A conservative position was adopted relative to name African elephants, which are assumed to be a single species.
∗ Sequences modified from [37]: tRNAs and control region added to cytochrome b.
Trois méthodes de construction d'arbres ont été utilisées : parcimonie non pondérée (MP), méthode du minimum d'évolution (ME) et maximum de vraisemblance (ML), les deux dernières méthodes étant fondées sur un modèle d'évolution de type TrN [27], dont les valeurs de paramètres ont été extraites du programme MODELTEST [28] à partir de l'analyse topologique d'un des deux arbres parcimonieux (pris de façon interchangeable).
Les résultats produits par les trois méthodes sont proches (Fig. 1). L'arbre le plus vraisemblable correspond à l'un des deux arbres parcimonieux, ces deux arbres ne différant que par la position d'un L. a. cyclotis (DRC2) dont la branche est très courte : ((DRC2, DRC9) DRC3) ou (DRC9 (DRC2, DRC3)). Ces phylogénies montrent (i) la monophylie des éléphants de forêt au sens large (cyclotis & pumilio) et (ii) une structuration nette des éléphants de forêt en deux clades (W pour « western », occidental, et E pour « eastern », oriental) qui ne recouvrent pas les limites systématiques en cours, ni une différenciation géographique absolue (du fait notamment de la présence de spécimens du Congo dans les deux clades). Contraindre la monophylie réciproque des cyclotis et des pumilio produit des arbres nettement moins parcimonieux et moins vraisemblables.
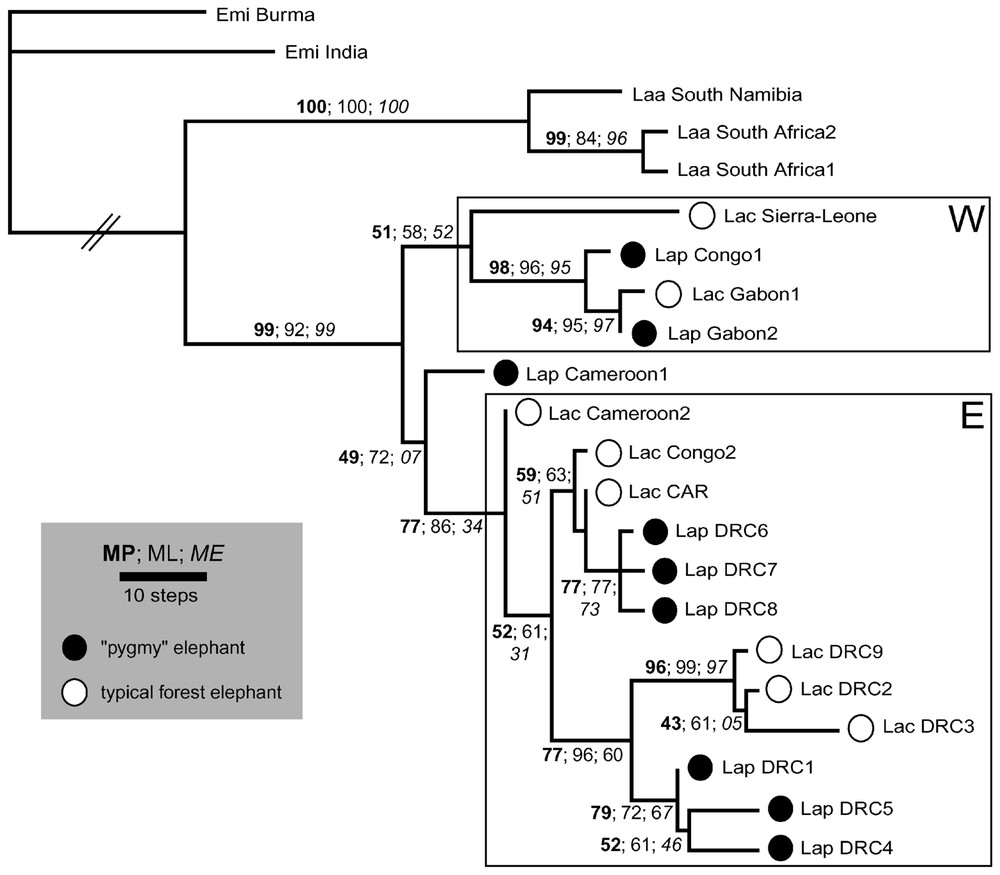
One of the two most parsimonious trees of MP analysis (282 steps; CIuninf = 0.66; RI = 0.8254; deltran optimisation). It is strictly identical to the ML tree (TrN model, see text for values). Bootstrap scores are displayed above branches for the three methods used.
Les deux spécimens du Cameroun disponibles dans notre échantillon ont un rôle à part dans cette phylogénie : ils proviennent de la zone de contact hypothétique entre les deux clades d'éléphants de forêt et nous aident à appréhender leur histoire commune. Le cyclotis (Cameroon 2), groupe frère des autres représentants du clade E, possède une branche terminale de longueur nulle, de sorte qu'il apparaı̂t comme un représentant de l'ancêtre moléculaire hypothétique de ce clade. Parallèlement, le pumilio (Cameroon 1) apparaı̂t comme un intermédiaire entre les deux sous-ensembles W et E. Comme le spécimen précédent, il repose sur une branche très courte de l'arbre, mais son assignation à l'un ou l'autre de ces clades est incertaine (et peu robuste) : la branche interne qui le rattache au clade E est extrêmement courte (Fig. 1) et, par des analyses de distance, il se classe comme groupe-frère du clade W. Ces résultats nous amènent à penser que la région du Cameroun pourrait appartenir au centre de dispersion primitif des éléphants de forêt en Afrique vers le bassin du Congo, d'une part, et vers les isolats forestiers occidentaux, d'autre part.
La cohérence géographique dans la répartition des haplotypes est frappante (Fig. 3) : les deux clades W et E possèdent des aires jointives, mais dont le recouvrement est faible. Et au sein de chacun de ces clades, la divergence entre les échantillons (pumilio et cyclotis) apparaı̂t être fonction de la distance géographique entre les taxons (Fig. 2). Ainsi, on retrouve des groupes monophylétiques régionaux où des spécimens pygmées sont mélangés à des cyclotis typiques.
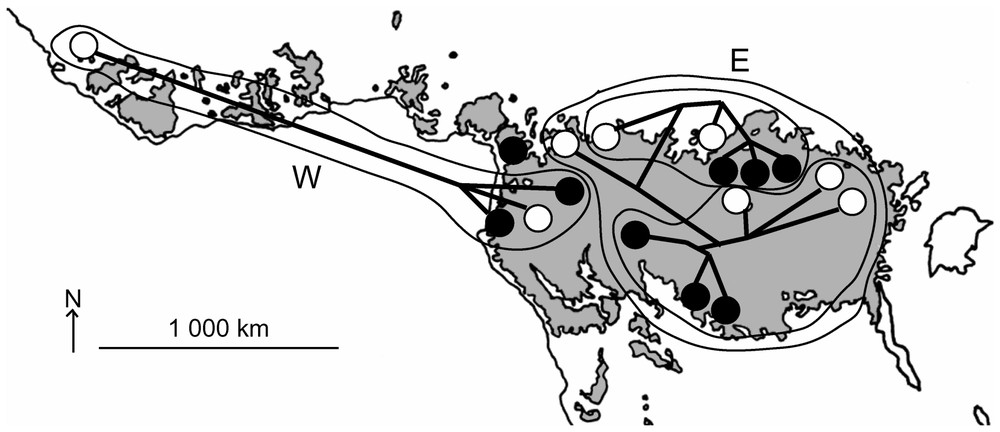
Geographical variation pattern within forest elephants. Forest patches of central Africa are displayed on a grey blackground. Major clades (W and E) are circled. As for Fig. 1, cyclotis and pumilio are depicted as white and black dots, respectively.
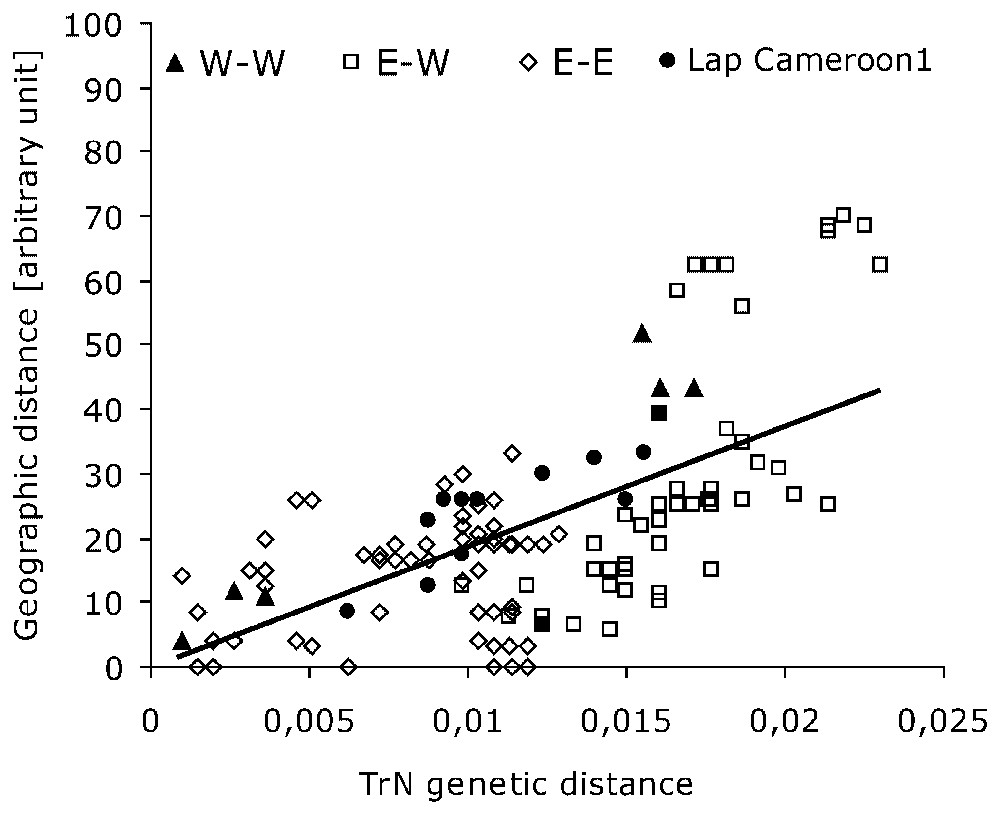
Genetic distance (TrN) plotted against rough geographical distance (km) for all specimens. r2=0.4045, r=0.6360, g=4.1749>2.575 for P=0.005.
Du point de vue des processus évolutifs, une seule hypothèse permet de rendre compte de façon satisfaisante de la répartition des données : il faut considérer qu'il n'existe pas de barrière reproductive entre les éléphants de forêt typiques et/ou pygmées. Dans ce registre, les différenciations locales entre les populations que nous observons ont pu s'établir par un phénomène d'isolement par la distance, associé à des événements historiques d'expansion et de rétraction des forêts en fonction des variations climatiques quaternaires. Dans ce cadre, il apparaı̂t nécessaire d'abandonner le taxon de rang spécifique Loxodonta pumilio. Il demeure toutefois possible que des populations locales d'éléphants de forêt aient évolué de façon convergente dans le sens d'une réduction globale de la taille. En somme, reconnaı̂tre que les éléphants pygmées ne constituent pas une espèce ne nie pas l'existence de tels animaux en tant que représentants d'un extremum local de variation au sein des éléphants de forêt.
1 Introduction
Recent years have seen the development of the molecular survey for the African elephant formerly recognized as a single species: Loxodonta africana (Blumenbach 1797) [1]. Since the publication of the first cytochrome b sequence of a forest elephant [2], molecular phylogenies have revived the debate over the systematic status of the forest form (subspecies) of the African elephant. The amount of genetic divergence (rather than the phylogenetic pattern) has been put forward to resurrect the view of two distinct species within African elephants: Loxodonta africana, the savannah elephant on the one hand, and Loxodonta cyclotis (Matschie 1900), the forest elephant on the other [3]. This position is consistent with the splitter-attitude of the early 20th century when the systematics of the African elephant was a permanent issue sustained by the promotion of new taxa out of the description of the most meagre morphological argument [4–7]. Of the 22 described taxa of African elephants, only three were retained for systematic discussions in the more recent literature: the savannah elephant (africana), the forest elephant (cyclotis), and the pygmy elephant (pumilio).
Until now, only two have been examined on molecular grounds: while both nuclear [3] and mitochondrial data [8] for cyclotis and africana forms have accumulated, nothing has been done concerning the so-called “pygmy” of central- and west-African rainforest: Loxodonta pumilio (Noack 1906). Loxodonta fransseni (Schouteden 1913) described as ‘l’éléphant d'eau' [9] was later synonymized to L. pumilio, but it is now obvious that none of these binomials is valid. The earlier was retained in the discussions on the African elephant, although the type specimen of L. pumilio – a captive specimen – appeared to be a young L. a. cyclotis that have continued growing until its death [18]. The subsequent binomial is held by a small-sized male specimen, but also juvenile – of D4/M1 dental aging (Debruyne's personal examination) – so that both can be considered nomina nuda. We keep the old species name, Loxodonta pumilio, just for communication and mainly for the sake of clarity, although the independence of the dwarf form is not supported in this article.
Throughout the last century, the existence of a dwarf form of the African elephant has been reportedly witnessed, although knowledge about it remained poorly documented [29] – mainly through captive specimens [10,11] –, and is always much debated. It was described as an extremely small form of forest elephant with shoulder height always inferior to 2 m, and rather comprised between 1.6 and 1.8 m [12–15]. Its geographical range is thought to encompass Equatorial Guinea, Gabon, Cameroon and Central African Republic (CAR), Congo and Democratic Republic of Congo (DRC, former Zaire) [16]. Its aquatic habits were also postulated [9,17,30]. For the latter reason, pygmy elephants have sometimes been regarded as a swamp-ecotype of the cyclotis form [22]. Besides, this remote habitat might have played a role in hindering research progress about this animal, which is mainly known through few collection specimens. The existence of pygmy elephants has also been denied [18,19] arguing they are nothing but juvenile forest elephants [20,21]; yet morphological evidence about the existence of dwarf-size mature elephants had been formerly attested [22]. Otherwise, it has been assumed that pygmy elephants are the results of individual cases of nanism or pathological growth [20]. As a compromise on this uncertainty, some authors assigned the pygmy elephants a subspecific rank: Loxodonta africana pumilio [17,23]. Interestingly, pygmy elephants seem to display the same level of morphological (discrimination through the size of adults) and ecological (distinct ecological preferendums) distinctiveness towards cyclotis as cyclotis does towards africana (but see [19] for competing point of view).
As the view of two species of African elephants is now fairly common [3,24–26], we question here the validity of the pumilio form as a third species. The issue of the status of pygmy elephants has been primarily sustained by anatomical studies [13,19,22] of dwarf-sized specimens in collection. We found it relevant to check the morphology of these historical specimens and analyse them on molecular grounds. We thus provide the first insight into the molecular phylogeny of nine collected specimens of dwarf loxodonts mainly derived from the Congo Basin, of which populations had not been investigated to date. The markers sequences are mitochondrial cytochrome b gene, and hyper-variable region 1 of the control region, plus in-between tRNAs.
2 Materials and methods
2.1 Sampling procedure, DNA extraction, amplification and sequencing
Two different types of samples were collected for our molecular purposes (Table 1). Hair (or tissues) were collected for three savannah elephants and two forest elephants. These samples were submitted to a standard CTAB extraction protocol [31]. Six bone samples from non-pygmy cyclotis individuals preserved in collection were also added. Samples of pygmy elephants – recognized as such through collection-labelling – were derived from nine specimens in collection. Among those preserved at the Royal Museum of Central Africa of Tervuren (MRAC, Belgium), five individuals (Nos. 7692, 9524, 20330, 20331, 20332) were already studied for morphology [22]. One is added (n°3396), representing the fransseni form of dwarf elephants from Mai-Ndombe Lake (former Leopold II Lake) area. Three other specimens are from the National Museum of Natural History of Paris (MNHN, France), which were all studied for their morphology [13,22]. Our observations confirm that these samples belong to extremely small individuals and at least five are adults (Nos. 1956–194, 3396, 7692, 9524, 20330, 20332). All bone samples were subjected to a modified phenol/chloroform protocol [32,33].
Until now 17 ‘forest elephants’ sensu lato have been examined (nine pygmies and eight ‘typical’ forest elephants). They broadly represent the geographic extent of forest elephant populations [34]. Three savannah African elephants are included in the analysis to test forest elephants' monophyly. Two Asian elephants are added as outgroup taxa, each one belonging to one acknowledged main subclade within Elephas maximus [35].
In this paper, we exclusively focus on the variability of mtDNA. This for two main reasons: (i) we consider here a majority of collection samples unlikely to have preserved high weight nucDNA and (ii) the higher level of variability of mtDNA demonstrated in previous studies of elephants [2] is expected to enhance topological resolution within African populations relative to nuclear markers [3]. We amplified and sequenced a highly variable fragment the mitochondrial chromosome spanning 1961 bp comprising sequences of cytochrome b gene (complete sequence, 1137 bp), threonine and proline tRNA genes (68 and 66 bp, respectively) and HVR1 of the control region (and central conserved region for a total of 690 bp).
Elephantid specific primers were designed with the entire mitochondrial sequence of Loxodonta africana [36] through oligo program (version 4.0) in order to sequence short overlapping fragments (see Table 2 for details). Long fragments (up to the entire length of the sequenced portion) could be obtained for samples of extant animals. Conversely, some of our bones fragments belonged to individuals preserved for more than 50 years in collections. For these specimens, the maximal fragment length that we could obtain at once was often within 400 bp. PCR and sequencing reactions were carried out with a protocol formerly published [37].
Set of primers for amplification and sequencing
| Primer | Primer sequence |
| L14096∗ | 5′-GCTTGATATGAAAAACCATCGTT-3′ |
| L14147 E∗ | 5′-ATGACCCACAYYCGAAAATCTCA-3′ |
| L14283 E∗ | 5′-TAACAGGATTATTCCTAGCCA-3′ |
| H14349 E∗ | 5′-TGGGATATAGATGAAAATGCA-3′ |
| L14421 E∗ | 5′-TCTGCCTATACACACACATTGGA-3′ |
| H14452 E∗ | 5′-GATGTTCCGTCCAATGTGTG-3′ |
| L14639 E∗ | 5′-TGAGGAGGCTTTTCRGTAGATAA-3′ |
| H14769 E∗ | 5′-GAATTGTTTGAGCCTGTTTCGTG-3′ |
| L14899 E∗ | 5′-AGACCCTGACCACTACATACC-3′ |
| H14946 E∗ | 5′-TGTAGGGGRGTATTTAGTGG-3′ |
| L14981 E∗ | 5′-GCCATCCTACGATCTGTACCA-3′ |
| H15038 E∗ | 5′-TTGATAGGAGTAGGGCTAGGA-3′ |
| L15151 E∗ | 5′-TACATGAATTGGCAGTCAACC-3′ |
| H15283 E∗ | 5′-TTACTTAATGAGGTAGTTTTCG-3′ |
| L15526 E | 5′-CGTGCATCACATTATTTACCC-3′ |
| H15593 E | 5′-GAATATGACTTGACACATTAGTTA-3′ |
| L15750 E | 5′-TACCTACCTCCGAGAAACTA-3′ |
| H15769 E | 5′-TGGTTTCTCGGAGGTAGGTA-3′ |
| H16127 E | 5′-TTATGTCCTCCGAGCATTGAC-3′ |
2.2 Sequence alignment and phylogenetic analyses
All sequences were read on both strands. Fragments obtained for collection samples were checked with blastn program [38]. Sequences were entered into a database using must package (version 2000 [39]). Semi-automated alignment was performed. No ambiguous positions were observed. Only one indel was found, in HVR1 portion, and it is distinctive for forest/savannah samples.
Phylogenetic analyses were performed using PAUP∗ (version 4.0b10 [40]). Unweighted Parsimony analysis (MP) was performed with the branch-and-bound algorithm of search. Any of the two shortest trees obtained was used as a topological reference in modeltest (version 3.06 [28]), to estimate the best-fitting model relevant to the data through the hLRT test, and yielded similar results. Maximum Likelihood (ML; 20 replicates of heuristic search with random sequence-addition and TBR branch-swapping algorithm) and Minimum Evolution (ME) analyses were conducted with the parameter values estimated for this model. The robustness of topologies was explored through bootstrap replications excluding uninformative characters: 1000 replicates for ME and MP, and 100 replicates for ML. Mantel test of correlation between genetic and geographic distances was performed with mantel non-parametric test calculator (version 2.0 [41]).
3 Results and discussion
3.1 Among congruence methods
The level of variability in the dataset is limited though four times greater than observed in nuclear intronic sequences available [3]. Two hundred sites are found variable, of which 148 are parsimony-informative (respectively 10.2 and 7.5% of the total sequenced fragment). The MP analysis yields two most-parsimonious trees of 282 steps that differ only in the internal branching within a three cyclotis clustering (DRC2 alternatively grouped with DRC3 or DRC9), due to extreme shortness of terminal branches. Homoplasy content of the dataset is reduced: CI excluding uninformative characters = 0.66; RI = 0.8254.
When any of the two parsimonious trees is used as topological reference in the modeltest procedure, the evolutionary model selected is TrN (for Tamura & Nei [27]), a moderate complexity model where both nucleotide frequencies (A=0.1895, G=0.0850, C=0.3817, T=0.3438), and three substitution rates are estimated from the data (two for the transitions: [AG] = 142.0750 and [CT] = 47.6429 relative to equal transversion rates fixed to 1.0000). Under these assumptions, ME and ML searches produce trees very similar to MP analysis: the most likely topology is shown in Fig. 1 and is strictly identical to one of the two most parsimonious trees, a situation that is not unexpected [42] and perhaps of general significance. The branch lengths displayed are those of deltran (MP) optimisation, but they are fairly consistent with those of TrN (ML) evolutionary model (data not shown). The ME tree displays slight differences in inner-branching for forest lineages, but these singularities are meaningless (no bootstrap score above 50). Reciprocally, all methods provide strong bootstrap support to the same nodes.
3.2 Phylogenetic pattern within forest elephants
The resulting pattern is clearcut (Fig. 1). The reciprocal monophyly of savannah elephants and ‘forest elephants’ (sensu lato) is retrieved and supported by the unique indel event in the matrix (optimised as a nucleotide loss within savannah lineage at site 1279, namely the 5′ end of the control region sequence). It conflicts with previous mitochondrial results [8] but seems along with nuclear data [3]. The monophyly of forest elephants is apparent due to the sampling of savannah elephants (southern Africa only). The question of the specific status of forest African elephants however is fully discussed elsewhere (Debruyne et al. in prep.).
Within ‘forest elephants’, two main clades are found. The first one comprises the most western samples (W), while the second is composed of mostly central and eastern individuals (E). Nevertheless, this division is not absolute insofar as Congo specimens are observed in both clades. Furthermore, its robustness is weak (in bootstrap proportions) and branch lengths information (plotted in Fig. 1) explains it. The cyclotis from Sierra-Leone indeed shows a long terminal branch in comparison with the internal branch of the subclade W, depleting his bootstrap score within this group. Conversely, the pumilio from Cameroon (Lap Cameroon 1), sister-taxon of all other subclade E members shows the shortest branch in the whole tree. Its assignation to any of the main clades is uncertain (and weakly supported): the internal branch that connects it to clade E is extremely short, and through distances analyses, it rather depicts a sister-taxon relationship toward clade W. Consequently, it appears very close to the putative ancestral haplotype of all ‘forest elephants’. Because it shares very few distinctive traits towards any of the other representatives of this lineage, it may be sensitive to random re-sampling of characters during bootstrap procedure, leading to uncertain alternate clustering with W or E group. Likewise, the cyclotis from Cameroon (Lac Cameroon 2) is the sister-taxon of other clade E specimens, and it shows a null terminal branch, so that it may represent the ‘molecular hypothetical ancestor’ of this clade. We face here a limitation of the tree representation for within-species phylogenetic investigations where both ancestral haplotypes and derived isolates can co-occur in populations [43]. Actually, these results suggest that the Cameroon area might belong to the primitive dispersion centre of forest African elephants into Congo basin on one side and into western forest isolates on the other.
With the exception of these former sequences, ‘forest elephants’ display a strong structure associated with high bootstrap values. Neither cyclotis nor pumilio form is found to be monophyletic and each appears in all main subclades of ‘forest elephants’. Some of them form local clades: DRC6 grouped with DRC7 and DRC8 for typical cyclotis, or the clustering of DRC9, DRC2 and DRC3 pygmy elephants. Nevertheless, enforcing topology to obtain monophyletic pumilio and/or cyclotis produces far worse likelihood scores (−lnL=4702.8931 for both as monophyletic or 4681.3908 and 4680.2329 for their respective monophyly, rather than 4485.2029 for the most likely topology), systematically rejected when compared to best tree by the S–H test [44].
3.3 Geographical distribution of the haplotypes
The observed phylogenetic pattern is at odds with the idea that two African forest elephants exist, but it prominently matches the regional distribution of our samples. Therefore, when a Mantel test of correlation between the genetic distance (estimated through TrN pairwise-distance matrix for cyclotis and pumilio) and geographic distance is performed among forest samples (Fig. 2), it is found highly significant (g=4.1749>critical value = 2.575, for P=0.005), so that there is a strong correlation between the genetic and geographic division in ‘forest’ elephants as a whole. The role of the most distant specimen (Lac Sierra-Leone) in this correlation has been examined: though lowering the global correlation score (rSL=0.4842 without Lac Sierra-Leone while r=0.6360 with it), its suppression does not preclude the significance of the Mantel test (gSL=4.7403). The pattern is consistent with the hypothesis of local isolation-by-distance of these elephants whatever their belonging to cyclotis or pumilio forms. It depicts the absence of reproductive barrier between these taxa, despite the remote and patchy nature of the forest areas they inhabit.
Likewise, when the phylogenetic relationships are superimposed over extant rainforest distribution (Fig. 3), the subclades W and E occupy distinct areas that may overlap in Cameroon/Congo. Strikingly, the two specimens from Cameroon display intermediate genotype: one of them (Lap Cameroon 1) shares few derived characters with either subclade W and subclade E, but also several autapomorphies, so that its relationships remain uncertain. The other one (Lac Cameroon 2) conversely shows no exclusive derived characters and clusters with all other subclade E specimens, as a potential representative of the ancestral genotype of this group. Cameroon samples also exemplify that both ‘pygmy’ and ‘non-pygmy’ elephants seem to share their habitat throughout their dispersion range.
The geographical division of our sample matches satisfyingly the refuge theory in tropical Africa [45]. Major climatic shifting has occurred in Africa for the beginning of the Quaternary. ‘Cold’ dry periods (when rainforest was depleted and hosted populations fragmented) succeeded to moist periods (when forest coalesced, up to achieving a continuum from Senegal to Uganda). These cyclic variations have surely disturbed forest elephant populations, which are known to rarely abandon the forest cover habitat. During dry periods, the most distant populations were surely apart and diverged from the others. Our data fit the hypothetic core areas where biodiversity is thought to have concentrated [45]: the most western populations are expected to be distantly linked to the Cameroon/Gabon area, which is itself apart from a Eastern DRC centre.
3.4 Potential explanations of the pattern
Two main phenomena may be responsible for the pattern we observe (between pumilio and cyclotis) at the specific-level or below. (i) Coalescence theory often accounts for the non-monophyletic patterns of specific taxa because of retention of ancestral polymorphism and subsequent lineage sorting. In the present case, the hypothesis that cyclotis and pumilio elephants behave as distinct reproductive units which share haplotypes inherited from a polymorphic ancestral population of ‘forest’ elephants is very unlikely. Indeed, the mean polymorphism (here the average gene diversity ) to be maintained through time in a lineage is highly dependent on the effective size (Ne) of this genetic unit (under the infinite-allele model, at the mutation drift equilibrium) [46]:
Yet each specimen of ‘forest’ elephant displays a unique haplotype, showing that our sampling procedure has been unable to account for the genetic diversity of extant forest elephant field populations. Although our sample displays a maximal within-diversity, it also betrays (despite not conforming a homogenous population) that we far underestimate the overall haplotype diversity. The effective size of the ancestral population of ‘forest’ elephants would have been huge for conveying such an amount of genetic variability. Moreover, subsequently to the putative speciation event, random lineage sorting would be responsible for the regional disruption in each taxa populations, but it cannot account for the geographical consistency that we observe between both pygmy and non-pygmy elephants (Fig. 3).
(ii) The alternate explanation is much simpler and convenient. It supposes that pumilio and cyclotis do behave as a single reproductive unit which populations have been isolated through time by distance and cyclic events of expansion and depletion of rainforest habitat. In that case, pygmy and non-pygmy elephants would share mitochondrial haplotypes because of reciprocal introgression.
4 Conclusion
Our data show that different lineages of dwarf elephants are apart in the central African populations tree and suggest a typical isolation-by-distance that promotes the position that ‘forest elephants’ as a whole behave like a single genetic unit. The recognition of pygmy elephants as a distinct species of African elephant is artificial. The relationships of the so-called pygmy elephants strongly advocates for abandoning the specific taxon L. pumilio or L. fransseni (whatever the precise status of the cyclotis form may be).
However, local genetic homogeneity of pygmy representatives should not be considered as a key argument to consider that pygmy elephants do not exist (that is to say that they are nothing but juvenile or pathological individuals): selection or drift may have promoted the dwarf size in remote elephant populations of central Africa. In that case, what was assumed a discrete pattern of variation (“dwarf or not dwarf”) may hinder a far more complex pattern of relationships at a populational level.
Acknowledgements
First and foremost we thank Dr. Wim van Neer and Wim Wendelen from the Musee Royal d'Afrique Centrale (Tervuren, Belgium), for sampling possibility and help on MRAC collection. Prof. Tranier, Dr. Cuisin, Prof. Robineau and Francis Renoult must be thanked for sampling facility in MNHN collections (Paris, France) as well as Michel Béaud for Fribourg Museum (Switzerland). We are also grateful to providers of present-day samples: ‘Parc zoologique de Vincennes’ (MNHN, France), ‘Parc zoologique de La Palmyre’ (France), Basel Zoo (Switzerland), Thoiry Reserve (France), and Ludovic Momont (Lope Reserve, Gabon). This study was supported by the ‘Service de systématique moléculaire’ of the MNHN. We thank Annie Tillier, Josie Lambourdière, Dr. Alexandre Hassanin for technical assistance, Dr. Nicolas Vidal, Dr. Sébastien Lavoué, Fabienne Rigal and Jacques Dubois for useful comments.



