1 Introduction
For nucleotide sequence data, DDBJ, EMBL and GenBank have collaborated for more than 17 years in constructing and serving as public databases, the International Nucleotide Sequence Databases (INSD). While INSD have operated under an excellent international collaboration, another important aspect was raised in research and development of biological databases, concerning gene expression data such as microarray, SAGE, EST and full-length cDNAs for which high-throughput technologies have recently been developed [1–4]. In particular, the accuracy of measurements with microarray technologies has substantially been improved and the spot density has tremendously been increased.
Under these circumstances, the first meeting for facilitating standardizations of microarray experimental data and their annotation was held by the Microarray Gene Expression Data (MGED) Group at EBI in 1999. Thereafter, the second MGED meeting was held also in Europe, and the third and fourth meetings were held in the United States. Those four meetings successfully attracted European and US researchers and engineers using microarrays. To extend the activity of MGED to Asian countries, we hosted the fifth MGED meeting in Japan in 2002. We believe that MGED has really been internationalized since the fifth MGED meeting.
MGED laid down and published standardizations for microarray experiments and data description called MIAME (Minimum Information About Microarray Experiment) [5]. MIAME has thus made it possible to construct a database of microarray data. In Japan, along with the MGED activity, we immediately started the development of a gene expression database in compliance with MIAME. This database is called CIBEX (Center for Information Biology gene EXpression database).
In this paper, we will outline CIBEX and propose international exchanges of gene expression data. CIBEX is also expected to play a key role in new research and development of comparative and functional genomics in the post-genome sequencing era.
2 Structure and design
The database management system (DBMS) of CIBEX is MySQL that operates under Linux. In this sense CIBEX is compatible with those that support the Japan Data Bank Center Database Management System (DBMS). The CIBEX DBMS is given in Fig. 1. In the DBMS there are basically relational tables which are intermingled with each other. Data submitted to CIBEX are automatically divided into pieces accordingly to those relational tables and distributed into the proper tables. Although we are aware of some demand of raw spot data to be stored and provided in a database, we do not accept raw spot data but authentically digitized one. This is mainly because raw data take up tremendous computer storage, which would be more and more serious as data are collected and accumulate in the future. However, it is possible to unidirectionally convert digital data to spot data in CIBEX.
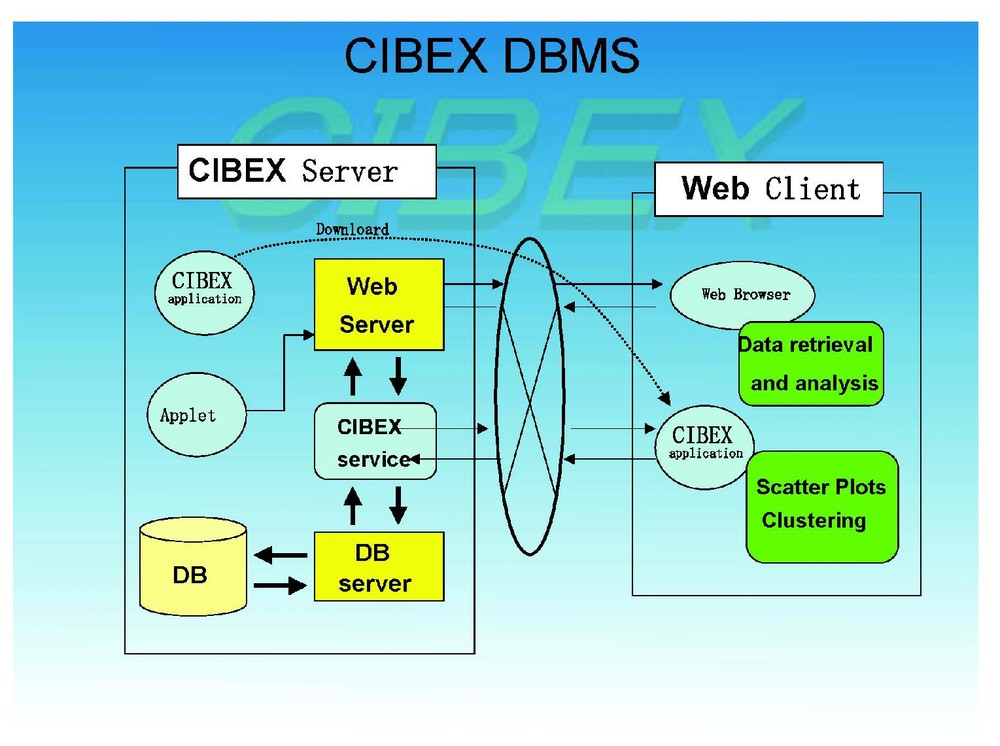
Outline of database management system of CIBEX.
3 Data submission
We have submission tools for data submitters to CIBEX through our web site. At present we have two submission tools, Stanford ScanAlize and GSILunomics ScanArray, which are in tab-delimited forms. Both formats are transferred into CIBEX through their respective loaders that are developed as Java applications. The tools each include items of gene symbol, gene name, the sequence accession number issued by INSD, spot locations (row and column), and measurements. Since our submission tools are in compliance with MIAME, the submitter unconsciously follows MIAME as long as he/she meets what each of our tools requires. To make the data submission more accurate and efficient between the data submitter and us, we first ask the data submitter to inform us of his/her contact information online. We then talk to each other online or on the phone about every bit of data submission. If everything is satisfied between the two parties, the submitter can then begin submission online in earnest. When the data submission is successfully completed, we issue a CIBEX ID number, by which the data is always handled during all processes in the CIBEX DBMS.
3.1 Data retrieval and visual viewer
CIBEX enables one to carry out data retrieval with respect to several data items such as CIBEX ID, experimental and biological conditions, gene names, authors, references, arrays, software and hardware as indicated in Fig. 2. If one retrieves data from CIBEX in terms of a particular experimental or a biological condition, one is led to the table information on the condition that shows gene manes, spot locations (row and column) expression intensities in number on a glass slide.

Manual page of CIBEX. The page shows the subject items for retrieval.
As mentioned above, CIBEX does not accept raw spots as input data, but it can convert digital data to spot images. This is because CIBEX is equipped with a user-friendly visual viewer that shows a set of spot images for the condition thus retrieved on the screen. One can then retrieve the information stored on each spot by clicking on the spot as shown in Fig 3. In this way one can easily know the gene name that is expressed, not expressed or equally expressed under particular experimental or biological conditions. One can also be informed about the intensity of expression of a gene through a histogram and line chart of each spot on the slide glass.

Graphical presentation of spot images. Converted spot images and their related pieces of information are shown.
For the set of spots retrieved one can obtain an image of scatter plot analysis with various X to Y ratios. From this image one is of course able to identify genes within the range of standard error and out of it. Combining the results of the expression intensity and of scatter plot analysis, one will be able to more confidently understand the expression status of a gene of interest. CIBEX also provides a tool for clustering analysis of the genes and samples in question.
3.2 CIBEX applications
The CIBEX service is made possible on Servelet container under Tomcat4 that includes the CIBEX service and JAVA servelet. The whole system works under Linux, as mentioned above. It is, however, compatible with Windows2000/XP or above. For launching applications CIBEX is equipped with JAVA Web Start. When one gets access to CIBEX for the first time, by use of JAVA Web Start one carries out download of the CIBEX service that at present includes scatter plot analysis, clustering analysis and visual viewer on the client server. Thereafter, one does not have to repeat download it again unless CIBEX updates or modifies its service. This makes the client server less burdensome and thus more efficiently interact with the CIBEX server.
3.3 Links
CIBEX has links with other useful databases such as INSD and PubMed. Therefore, if a retrieved spot contains cDNAs that have been registered at INSD with their accession numbers, one can visit INSD with the accession numbers and get access to the information on them there. Since CIBEX also enables one to retrieve against it with respect to references, one can immediately surf the net to PubMed to obtain more detailed information about the experiment or gene in question. In this way one can extend knowledge about gene expression to DNA and protein sequences to their homologs and to the related literature.
4 Future plans
Since our prime objective for developing CIBEX is to collect expression data from researchers mainly from Asian countries and distribute the data worldwide, we will contact and stimulate them to submit their data to us. To facilitate the data collection, we also began to collaborate with ArrayExpress at EBI [6], to which the authors who submit their papers containing the original microarray data to a few journals (Nature [7], Science and Lancet) are required to submit their data. The first step of the collaboration is to exchange data between CIBEX and ArrayExpress on the basis of MAGE-ML. The collaboration in data exchange will be extended to GEO at NCBI [8]. In this way we will be able to establish the international collaboration in data exchange among CIBEX, ArrayExpress and GEO.
An important aspect in data collection is to provide the data submitters with something beneficial. We think that the most beneficial one is to enable researchers in the world to compare expression data produced by different laboratories. There are two major ways to create this environment; one is to establish the international collaboration mentioned above, and the other is to make CIBEX more useful by developing data retrieval and analysis tools. We believe that this is one of the main objectives of the MGED Society.


