Version française abrégée
La protéine 4.1R (ou 4.1R) est l'un des composants clés du squelette érythrocytaire, dont le rôle majeur est de stabiliser le complexe spectrine/actine, via son domaine interne de 10 kDa (domaine SAB). La déficience en cette protéine se traduit par une anomalie morphologique du globule rouge, l'elliptocytose héréditaire 4.1R(−). Cette anomalie est asymptomatique à l'état hétérozygote, mais elle se traduit en cas d'absence totale de protéine par une anémie hémolytique grave due à une fragilisation accrue de la membrane, et nécessitant des transfusions répétées.
4.1R est codée par un gène unique de plus de 200 kb. L'isoforme érythrocytaire de 80 kDa est le prototype d'une famille d'isoformes de tailles variant de 30 à 120 kDa. Ces isoformes proviennent dans leur grande majorité de multiples épissages différentiels du pré-messager, et leur expression dépend du type de tissu et du stade de différenciation. L'isoforme érythrocytaire est de loin la mieux connue ; elle contient quatre domaines structuraux définis suite à une digestion ménagée : N–30 kDa–16 kDa–10 kDa–22/24 kDa–C.
Dans un précédent travail, nous avions analysé les isoformes 4.1R dans des tissus perfusés de rat et de souris, focalisant sur la région codant le domaine SAB. Ce travail révéla que le muscle squelettique exprime, de façon presque exclusive, un nouvel exon alternatif, l'exon 17a. Nous avons alors émis l'hypothèse que cet événement d'épissage devrait contribuer à la production d'une isoforme dotée d'un rôle physiologique particulier, et qui contient un peptide spécifique du muscle (MSP : muscle-specific peptide) codé par l'exon 17a. Plus tard, il a été établi que 4.1R, présente sous deux formes majeures de 105/110 kDa et 135 kDa, est exprimée au niveau des bandes A du sarcomère, et s'associe aux protéines du système contractile, myosine, actine et tropomyosine via son domaine de 10 kDa.
Le travail présenté ici repose sur la production d'un nouvel anticorps dirigé contre MSP, et dont les tests en Western Blot ont prouvé la spécificité. Il reconnaît les protéines 4.1R(+MSP), aussi bien chez l'homme que chez la souris. Cet anticorps a été d'abord utilisé pour marquer des coupes transversales de muscle normal ou de muscle atteint de myopathie de Duchenne (DMD). Ces expériences ont révélé une expression au niveau du sarcolemme de muscle normal, laquelle expression disparaît dans le muscle déficient en dystrophine. Cette observation suggère que l'assemblage de la protéine 4.1R au sarcolemme est dépendant de la dystrophine. On sait depuis plusieurs années que l'absence de dystrophine dans le muscle myopathique entraîne en effet des déficiences secondaires en d'autres composants du sarcolemme, telles les sarcoglycanes. En appui à cette observation, un double marquage avec des anticorps anti-4.1R(+MSP) et anti-dystrophine a révélé une expression partielle de 4.1R, colocalisée avec la dystrophine résiduelle, exprimée au niveau du sarcolemme de muscle atteint de myopathie de Becker (BMD).
Le sarcolemme exprime aussi la β-spectrine, le ligand de 4.1R au niveau du squelette érythrocytaire. Cependant, il est probable que les isoformes musculaires de ces deux protéines ne s'associent pas dans le muscle. En effet, la déficience en dystrophine dans le muscle DMD entraîne une déficience en 4.1R, sans affecter l'expression de β-spectrine au niveau du sarcolemme.
En plus de l'expression de 4.1R au niveau du sarcolemme, les expériences d'immunohistochimie ont révélé un marquage au niveau du myoplasme des fibres, aussi bien dans le muscle normal que dans le muscle myopathique. Un double marquage utilisant des anticorps spécifiques des types de fibres a permis de démontrer que les isoformes 4.1R(+MSP) sont préférentiellement exprimées dans les fibres rapides.
L'anticorps anti-4.1R(+MSP) a été ensuite utilisé pour identifier les isoformes protéiques par Western Blot. Ces expériences ont montré que les isoformes contenant le peptide musculaire MSP apparaissent sous forme d'un doublet d'environ 96/98 kDa. Ce dédoublement rappelle la forme dédoublée du squelette érythrocytaire de 78/80 kDa, laquelle correspond à une déamidation d'un résidu asparagyl. Une analyse par Western Blot au cours d'une cinétique de différenciation des myoblastes C2C12 a permis de montrer que les isoformes 4.1R(+MSP) ne sont exprimées ni dans les myoblastes ni dans les myotubes en formation, et que leur apparition est propre au tissu musculaire différencié.
Pour identifier de manière exhaustive les isoformes de 4.1R contenant l'exon 17a, nous avons amplifié les messagers 4.1R par RT-PCR, utilisant deux couples d'amorces. Afin d'amplifier spécifiquement les molécules contenant l'exon 17a, nous avons choisi d'utiliser dans chacune des deux séries de RT-PCR une amorce dans cet exon. Ces expériences ont permis d'identifier deux isoformes majoritaires, l'une contenant l'exon 17a entier, l'autre la forme tronquée 17a′. Ces isoformes majoritaires contenaient par ailleurs tous les exons, à l'exception des exons 3, 14, 15 et 17b. Deux formes minoritaires ont été aussi identifiées, l'une et l'autre contiennent en plus l'exon 17b et diffèrent par la présence, soit de l'exon 17a entier soit sa forme tronquée 17a′.
Pour savoir si l'expression tardive des isoformes protéiques résulte d'une régulation au niveau du métabolisme des ARN messagers, nous avons entrepris une analyse du profil d'épissage des exons codant le domaine SAB au cours de la différenciation des myoblastes C2C12 et dans le muscle mature. Ce travail conclut à l'apparition asynchrone des exons alternatifs 16 et 17a. En effet, les isoformes majoritaires exprimées dans les myoblastes individualisés ne contiennent ni l'un ni l'autre de ces deux exons. La fusion des myoblastes en myotubes est accompagnée par l'apparition de l'exon 16 seul dans les isoformes majoritaires. L'apparition de l'exon 17a est en revanche nettement plus tardive, puisque les isoformes contenant cet exon ne deviennent majoritaires que dans le tissu différencié. Ces résultats sont en accord avec l'observation, rapportée plus haut, d'une expression tardive des isoformes protéiques contenant le peptide MSP.
1 Introduction
The 80-kDa isoform of protein 4.1R promotes binding of spectrin and actin, two major proteins of the cortical skeleton at the red cell membrane [1]. Four structural domains of human erythrocyte protein 4.1R have been deduced from limited proteolytic and chemical degradation: 30, 16, 10 kDa, and 22/24 kDa from N-terminal to C-terminal [2]. The activity needed for the spectrin/actin complex formation resides in the 10-kDa domain [3]. In fact, the prototype 80-kDa protein 4.1R is a member of a large family of immunoreactive polypeptides, ranging from 30 to 210 kDa in size, and found in a large array of normal tissues [4,5]. Most of these isoforms are generated by alternative splicing of 4.1R pre-mRNA.
4.1R pre-mRNA is encoded by a single gene [6,7], EPB41 gene, but undergoes a highly regulated alternative splicing involving many sequence motifs [7–11]. Two particular sequence motifs significantly direct the functional behaviour of protein 4.1R in red cells: (i) a 17nt motif at the 5′ end of exon 2 (5′E2), the exclusion of which removes the upstream AUG1 translation initiation codon, allowing the use of the downstream AUG2 initiation site (Fig. 1A); and (ii) exon 16, the inclusion of which leads to the production of a functional 10-kDa spectrin/actin binding (SAB) domain. These two splicing events are regulated in an asynchronous fashion during erythroid differentiation [12]. It has also been documented that splicing regulation of both 5′E2 and exon 16 is functionally crucial for targeting protein 4.1R isoforms to cell compartments in nucleated cells [13–17].
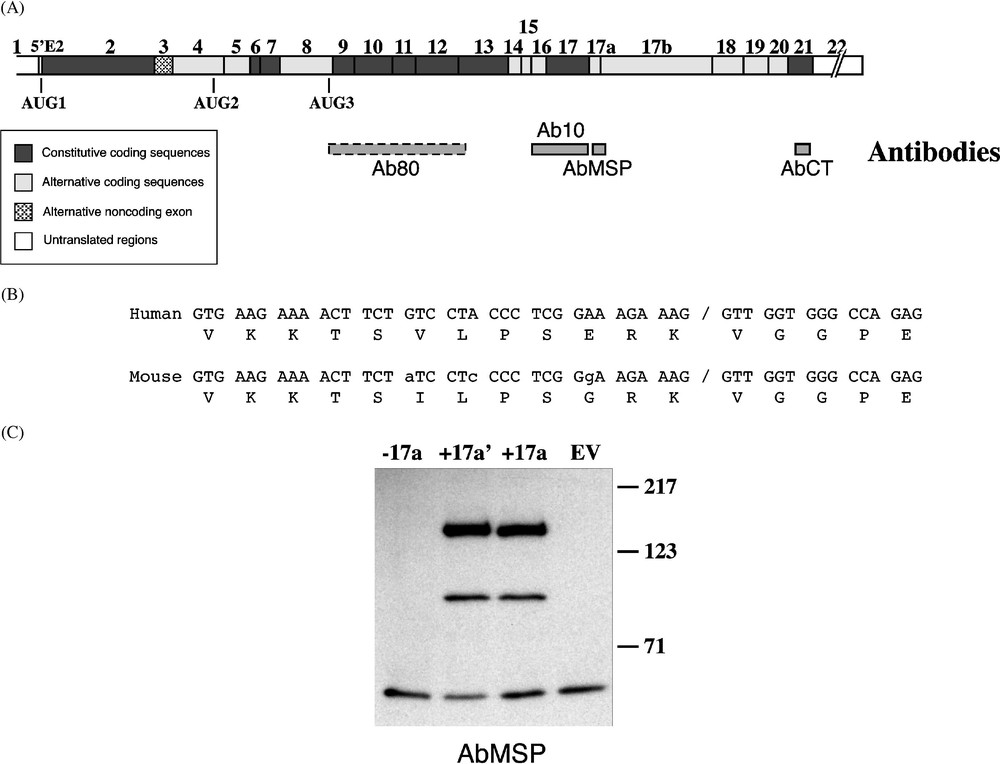
4.1R antibodies. (A) Epitopes of antibodies against the whole 80-kDa erythroid isoform (Ab80; this antibody is represented with a dashed box because its epitopes are unknown), the 10-kDa SAB domain (Ab10), the MSP (AbMSP), and exon 21-encoding C-terminal peptide (AbCT), are schematically depicted under a composite cDNA diagram of 4.1R. (B) Sequence alignment of exon 17a in mouse and human. Nucleotide mismatches are indicated in small letters in the mouse sequence. Of note is the peptide sequence similarity; among the 17 residues, only two positions are divergent between the two species. (C) Characterization of AbMSP. Three cDNA constructs served for in vitro translation of 4.1R protein isoforms. (−17a) lacks exon 17a; contains the short form of exon 17a; (+17a) contains the full-length exon 17a. EV: empty vector. Western blot analysis using AbMSP revealed specifically 4.1R isoforms containing either exon 17a′or 17a.
Analysis of 4.1R mRNA splicing pattern in various perfused tissues has revealed the presence of a unique sequence motif virtually confined to skeletal muscle [7]. This motif is encoded by an individual exon, exon 17a, and appears as a doublet upon RT-PCR amplification, which corresponds to the inclusion of either the whole 51-nucleotide exon, or the 36nt at the 5′ end, namely exon 17a′ [7]. The latter mRNA species is most likely generated by alternative splicing using an internal donor splice site. In all cases, the alternative splicing does not alter the translation reading frame. We therefore hypothesized that the muscle-specific 17 amino acid peptide (MSP) encoded by exon 17a must be part of a protein isoform with specific functional properties in muscle. In total, the region of 4.1R mRNA encoding the SAB domain actually includes three alternatively spliced exons, namely exons 14, 15, 16, and separated from the C-terminal encoding domain by two additional alternative exons, exons 17a and 17b (Fig. 1A).
A recent work has used immunofluorescence microscopy to study the intracellular distribution of 4.1R in longitudinal sections of skeletal myofibres [18]. This study has shown that 4.1R colocalizes with the thick myosin filaments at the A-bands. It further showed that 4.1R co-immunoprecipitates with three major contractile proteins, α-actin, myosin, and tropomyosin, hence demonstrating that 4.1R is an intrinsic component of the myoplasm.
In the present study, we raised an antibody against the MSP, and revealed upon immunohistochemical analyses that the MSP-containing isoforms are preferentially expressed in the myoplasm of fast myofibres of differentiated muscle. In addition, we documented the presence of 4.1R epitopes at the sarcolemma of normal skeletal muscle. In muscle from patients with Duchenne muscular dystrophy (DMD), or Becker muscular dystrophy (BMD), the fibre type-specific staining of the myoplasm were similar to those of normal muscle. By contrast, we found a 4.1R deficiency along with the dystrophin deficiency in the sarcolemma. We finally examined the alternative splicing pattern of exons around the muscle specific exon. We found that exons 16 and 17a are predominantly skipped in myoblasts, but subsequently incorporated in an asynchronous fashion.
2 Materials and methods
2.1 Cell culture and induction of differentiation
C2C12 cell line obtained from the American Type Culture Collection (Rockville, MD, USA), was maintained in non confluent culture as distinct mononucleated myoblasts in DMEM, supplemented with 4.5 g l−1 glucose and 10% FCS (Life Technologies, Gaithersburg, MD, USA), a medium recommended for cell proliferation. To promote muscle differentiation, confluent cells were cultured in (DMEM, 4.5 g l−1 glucose, 0.5% FCS). Proliferating myoblasts and forming myotubes were washed with 1X PBS, trypsinised, and harvested for RNA and protein isolation.
2.2 mRNA analysis
Total RNA from muscle tissues or cultured cells was extracted following cell lysis by ultracentrifugation through a CsCl step-gradient as previously described [7,12], or an alternative procedure using the RNeasy Total RNA kit (Qiagen, Hilden, Germany). RT-PCR experiments were performed on 1 μg of total RNA as previously detailed [7,19]. PCR fragments were subcloned into pSTBlue-1 (Novagen, Madison, WI, USA) vector, and sequenced using SP6 and T7 primers.
2.3 Primary antibodies
Monoclonal antibodies against dystrophin (AbDYS2), β-spectrin (AbSPEC2), fast (AbMHCf) and slow (AbMHCs) type myosin heavy chains, were purchased from Novocastra Laboratories (Newcastle, UK). Anti-laminin polyclonal antibodies were obtained from Sigma Immunochemicals (Saint Louis, Missouri). Affinity-purified polyclonal antibodies directed against the 4.1R 80-kDa erythroid isoform (Ab80), the 10-kDa spectrin–actin binding domain (Ab10), and a peptide encoded from exon 21 at the C-terminus (AbCT) were all described and used previously [19,20]. Monoclonal antibody C18 (AbC18), immunologically reactive against the 22–24 kDa C-terminal domain ([21] and data not shown) was kindly provided by Dr M. Garbarz. A polyclonal antibody, AbMSP, was raised in rabbits against the predicted sequence of the MSP [NH2-KKTSVLPSERKVGG-CONH2] [7]. This peptide was synthesized by Synt:em (Nîmes, France), and the antibody was obtained from Research Genetics (Huntsville, Alabama). Peptide coupling, immunisation protocol, and affinity purification were as previously described [19].
2.4 Isolation and immunoblotting analysis of muscle proteins
Liquid nitrogen quick-frozen muscle biopsies were resuspended in a solution containing 75 mM Tris-HCl, 10% SDS, 10% glycerol, 5% β-mercaptoethanol, 5% protease inhibitors Complete™ (Roche, Mannheim, Germany), and bromophenol blue. C2C12 mononucleated individual cells (), and myotubes were recovered from culture flasks following trypsin treatment, pelletted, and resuspended in 75 mM Tris-HCl, 15% SDS, 20% glycerol, and bromophenol blue.
Protein samples (∼300–500 μg) were separated by SDS-PAGE on 6% acrylamide gels, transferred electrophoretically onto nitrocellulose membrane. Transferred proteins were visualized by soaking the membranes in Ponceau Red (Sigma Chemical Company) for 5 min at room temperature. The staining was washed out by soaking the membranes twice (10 min each) in distilled water. Membranes were blocked by incubation for 1 h in TBS containing 10% non-fat dry milk, and were then washed three times (10 min each) with TBS containing 0.25% Tween 20. Following an incubation with polyclonal or monoclonal primary antibodies in TBS/0.1% Tween 20 overnight at 4 °C or 2H at room temperature, the membranes were washed three times with TBS/0.25% Tween 20, and incubated with horseradish peroxidase-labelled secondary antibody (Amersham Pharmacia Biotech Europe, Orsay, France), 1:10 000 diluted in TBS/0.1% Tween 20, for 1 h at room temperature, then washed three times with TBS/0.25% Tween 20. Immunoreactive bands were detected using the ECL+ Western blotting detection system (Amersham Pharmacia Biotech), according to the manufacturer's recommendations.
2.5 cDNA constructs and in vitro transcription and translation
Three 4.1R cDNA fragments were prepared following RT-PCR amplification of human RNA templates obtained from MOLT4 or skeletal muscle. The DNA fragments were then cloned into pGEM-4Z vector (Promega, Madison, WI, USA). Selected clones, namely (), (), and () were sequenced prior to use in in vitro experiments. They all contained the three in-frame AUGs (Fig. 1A). The three clones were identical, except that () was lacking exon 17a, and () and () contained the alternative form 17a′ or the full-length exon 17a [7], respectively.
In vitro transcription/translation of 4.1R was performed using the TNT-T7 coupled reticulocyte System (Promega), in the presence of 35S-methionine. Newly synthesized proteins were authenticated, and their quality controlled by autoradiography following electrophoretic separation on 8% SDS-PAGE. The transcription/translation step was then repeated in the presence of non-radioactive methionine, and the generated proteins were analysed by Western blot using AbMSP and revealed by the ECL+ detection procedure.
2.6 Immunofluorescent confocal microscopy
Transverse cryosections, 7 μm thick, of normal DMD or BMD human skeletal muscle were prepared on a cryostat from muscle biopsies. Muscle samples were first blocked for 1 h at room temperature with phosphate buffered saline (PBS) containing 10% bovine serum albumin and 0.1% Tween 20. They were then incubated with primary antibodies in PBS containing 0.1% Tween 20, for 2 h. After thorough washing with PBS, 0.1% Tween 20, the samples were incubated for 1 h with the appropriate secondary antibodies: goat anti-rabbit Ig's conjugated to fluorescein isothiocyanate (BioSource International, Camarillo, CA) or Alexa 488 (Molecular Probes, Eugene, OR) and sheep anti-mouse Ig's Texas Red linked (Amersham Pharmacia Biotech AB, Uppsala, Sweden) or Alexa 546 (Molecular Probes), diluted in PBS, 0.1%Tween 20 according to the suppliers' recommendations. The samples were finally washed three times in PBS, and mounted with Fluoprep (BioMérieux, Marcy-l'Étoile, France) or with Tris HCl 150 mM, glycerol 25%, pH 8.6. When needed, muscle sections were stained with 4′,6′-diaminophenylindole (DAPI) for 5 min, and rinsed briefly in PBS before mounting. Double immunofluorescence staining was performed by mixing the secondary antibodies. All the incubations were performed at room temperature in a humidified chamber. The samples were observed with a confocal Zeiss Axiovert 135 microscope through a 40× oil immersion objective. Images were processed using Photoshop software (Adobe systems, Inc., San Jose, CA).
3 Results
Both exon 17a and its shorter form 17a′ were found in human and mouse [7]. Sequence alignment revealed a high degree of identity between these species (Fig. 1B). In particular, the sequence at and surrounding the cryptic 5′ splice site within the exon is identical, suggesting that activation of this cryptic site might have a physiologic relevance. The encoded protein sequence diverges between the two species at two residues (Fig. 1B). To identify MSP-bearing 4.1R protein isoform(s) in muscle, a new antibody, AbMSP, was specifically raised against the MSP. We first ascertained its reactivity with both human and mouse muscle tissues (data not shown). We then tested its specificity by Western blot using 4.1R proteins, in vitro translated from cDNA constructs that lack exon 17a (), or contain either the alternate exon 17a′ () or the whole exon 17a (). As shown in Fig. 1C, AbMSP revealed the two expected isoforms initiated at AUG1 and AUG2, that were exclusively generated from () and () constructs. These isoforms are identical to the well-identified 135 and 80 kDa isoforms [8,9], except that the latters are missing the MSP sequence motif. The antibody did not cross-react with the isoforms generated from construct or from the empty vector (EV). These data suggest that AbMSP is specific to isoforms that contain either of the peptides encoded by 17a′ or 17a.
3.1 Protein 4.1R is targeted to normal but not to DMD sarcolemma
Immunohistochemical experiments were performed to investigate 4.1R expression in muscle. In control muscle samples, a consistent and distinct staining at the sarcolemma was observed. Higher magnification ascertained that the staining is localized at the sarcolemma and not the interfibre connective tissue (Fig. 2A–D). Similar results were obtained with the antibodies Ab80, AbCT, and AbC18 (not shown). Moreover, a spot-like staining was also observed in the spaces between the myofibres (Fig. 2B). Using DAPI antibodies to label nuclear DNA, double immunohistochemical experiments showed that this punctate labelling does not colocalize with the nuclei (not shown). However, it colocalized with vessel sections in double immunostaining with anti-laminin that served to label vessel walls (not shown). These data suggest that 4.1R that appears between the fibres most likely corresponds to blood cell 4.1R rather than to a muscle 4.1R component per se. In all these experiments, no significant staining was obtained in a control set of sections using the secondary antibodies alone, suggesting that the labelling was specifically generated by 4.1R antibodies.
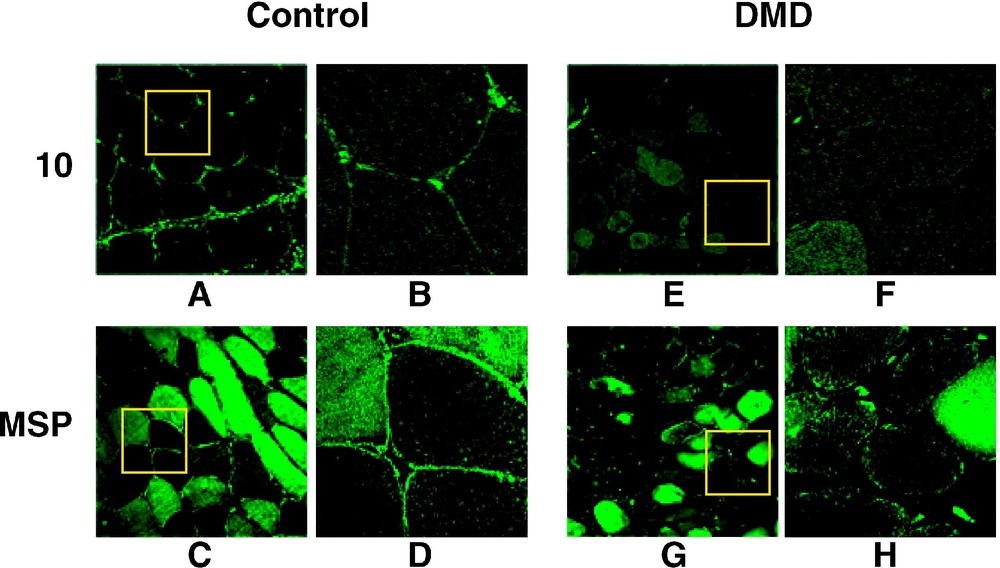

4.1(+MSP) isoform in present in normal but not in DMD sarcolemma. (A–H). 4.1R antibodies Ab10 (10) and AbMSP (MSP) were used to label control and DMD muscle cross-sections. The antibodies stain the sarcolemma in normal but not in affected muscles. For each anti-4.1R panel, the right view (B, D, F, H) is a magnification of the inset square. (I–L) Immunolabelling with anti-dystrophin (Dys) and anti-β-spectrin (Sp) are shown to emphasize the β-spectrin assembly to the sarcolemma by contrast with the complete absence of dystrophin in DMD muscle.
The same set of antibodies was also used to examine the sarcolemmal 4.1R expression in muscle biopsies obtained from DMD patients. In all cases, 4.1R epitopes were absent from the sarcolemma (Fig. 2E–H). Knowing that DMD muscle sarcolemma completely lacks dystrophin, but expresses β-spectrin, we used these markers to ascertain a proper immunolabelling of the sarcolemma. As shown in Fig. 2I–L, a specific antibody against dystrophin labelled the control, but not the DMD muscle sarcolemma, whereas anti-spectrin antibody stained both control and DMD muscle cells. To further support these observations, muscle cross sections obtained from BMD patients were double labelled with AbMSP and AbDys antibodies. As shown for BMD45-55 in Fig. 3, 4.1R epitopes colocalize at the sarcolemma with partial expression of dystrophin. Altogether, these data suggest that 4.1R distribution at the sarcolemma coincide with that of dystrophin.
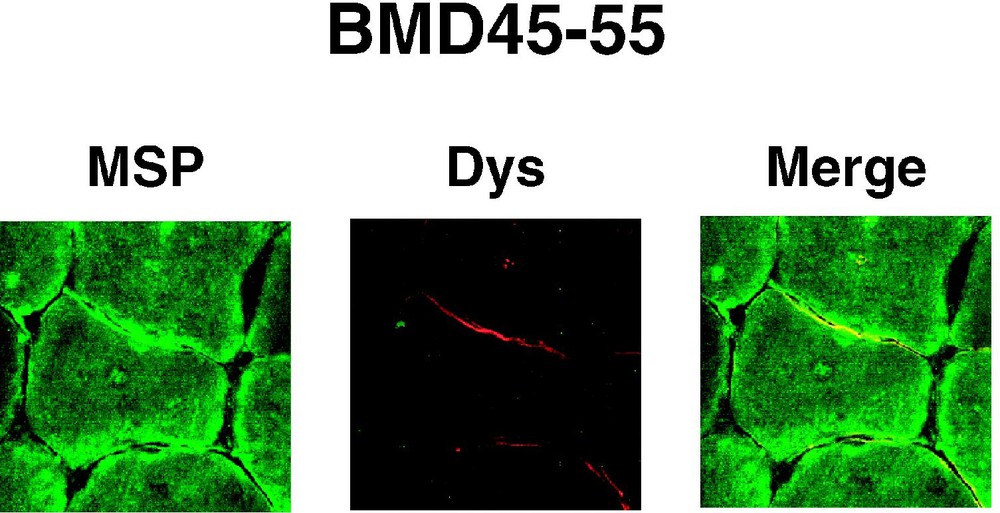
4.1(+MSP) isoform colocalizes with dystrophin in BMD sarcolemma. Muscle cross sections obtained from a patient with the BMD45-55 deletion of the dystrophin, were double labelled with AbMSP (MSP) and AbDys (Dys) antibodies. Partial expression of dystrophin colocalizes with partial expression of 4.1R at the sarcolemma.
3.2 Protein 4.1R is targeted to fast myofibres in normal and DMD muscle
In addition to the sarcolemmal staining, immunohistochemical experiments revealed a distinct labelling of the fibre myoplasm. However, this myoplasm staining appeared heterogeneously distributed, particularly with AbMSP (Fig. 2C). This observation suggested a protein expression or abundance related to myofibre types. To test this hypothesis, we subsequently performed a co-immunofluorescent staining using AbMSP and fibre type-specific antibodies (Fig. 4). Muscle fibre types were histochemically defined using fibre-specific anti-myosin heavy chain (MHC) antibodies: AbMHCf and AbMHCs for fast and slow myofibres, respectively (Fig. 4A and G). AbMSP generated a strong staining that coincided with the signal obtained with AbMHCf (Fig. 4A, C and E). On the other hand, a complementary labelling was obtained using AbMHCs and AbMSP (Fig. 4G, I and K). Absence of primary antibodies showed no significant labelling (not shown). In DMD muscle cross sections, AbMSP revealed a pattern similar to that observed in normal muscle sections: immunofluorescence staining coincides with the fast myofibre types on one hand (Fig. 4B, D and F), and complements the slow fibre labelling with AbMHCs, on the other hand (Fig. 4H, J, and L). Ab10 was also used in double immunofluorescence experiments and turned to also label the fast myofibres (not shown).

4.1(+MSP) isoform is present in both normal and DMD fast fibre myoplasm. AbMSP was used in co-immunostaining with either fast myofibre-(MHCf) or slow myofibre-(MHCs) specific antibodies. AbMSP labelled specifically the myoplasm of the fast myofibres in both normal (A, C, E) and DMD (B, D, F) muscles. Note the complementary staining generated by AbMSP and MHCs (G–L). In addition to the myoplasm, AbMSP revealed a distinct staining at the sarcolemma of all fibres in normal but not in DMD muscle.
In all these experiments, while all normal myofibres were immunoreactive to either of the two anti-myosin antibodies, part of the DMD myofibres, corresponding probably to degenerating fibres, responded neither to any of the 4.1R antibodies nor to myosin type-specific antibodies.
Altogether, these data suggest that protein 4.1R is targeted to fast myofibre myoplasm.
3.3 A major 4.1R protein isoform is expressed in muscle
4.1R protein expression was next analysed by Western blot in human (not shown) and mouse muscle biopsies, as well as in myoblasts, and during myotube formation of C2C12 cultured cells (Fig. 5). AbMSP revealed a predominant and consistent band, henceforward named 4.1R(+MSP), that appears as a doublet at ∼ 96/98 kDa, exclusively in differentiated muscle. A previous work has also described a 4.1R doublet in skeletal muscle, but the estimated size was about ∼ 105/110 kDa [18]. In addition to these bands, minor isoforms of higher molecular weights (∼135 and apparent molecular weights) were revealed. They probably result from alternative splicing pathways that lead to a translation initiation at the upstream AUG1 (see Fig. 1A). To confirm this observation, Western blot was performed using Ab10, an antibody that recognizes the flanking peptide sequence upstream of MSP, and which corresponds to the SAB domain in red cell membrane 4.1R isoform. The immunoblot shown in Fig. 5 (right panel), displays a pattern very similar to the expression pattern described above. Again, the major isoform is completely absent from the myoblasts and differentiating myotubes. The occurrence of 4.1R(+MSP) isoform as a doublet recalls the erythroid 4.1R doublet, which derives from the deamidation of an Asn residue [22]. Minor bands of low molecular weights appear using either of the antibodies AbMSP or Ab10 (Fig. 5). The identity of these bands remains unknown.
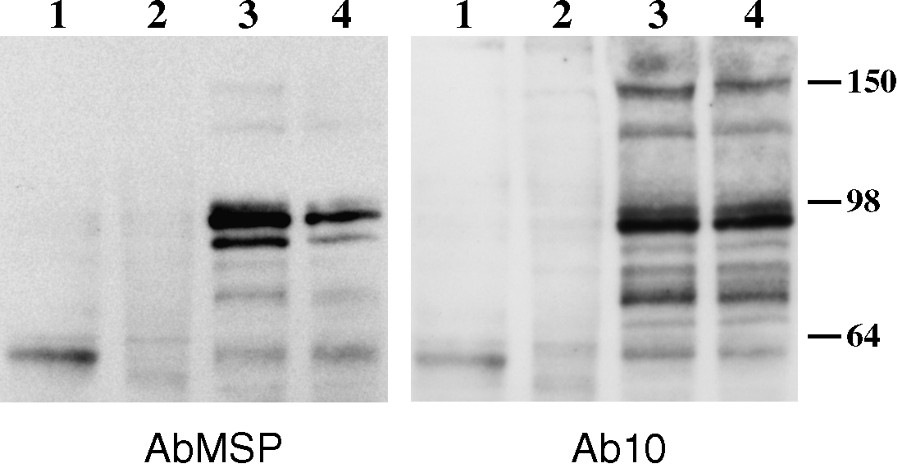
Protein 4.1R(+MSP) isoform expression in skeletal muscle. 4.1R expression in skeletal muscle and during muscle differentiation. Western blot analysis was performed, using AbMSP antibody (left panel), or Ab10 antibody (right panel), on protein homogenates obtained from cultured myoblasts (lanes 1), myotubes (lanes 2), and mature mouse muscles gastrocnemius (lanes 3), and soleus (lanes 4) About 450 μg of proteins was loaded. Size markers in kDa are indicated.
Collectively, these results demonstrate that (i) skeletal muscle expresses a major isoform of and minor isoforms of higher molecular weights, and that (ii) these isoforms contain the MSP, encoded by exon 17a, along with the SAB domain encoded by the flanking exons 16 and 17.
3.4 Multiple alternative splicing events of 4.1R pre-mRNA occur during muscle differentiation
Occurrence of MSP-containing protein isoforms only in differentiated muscle suggested a late expression of these isoforms. This observation prompted us to examine 4.1R mRNA splicing pattern during muscle differentiation.
Total RNA from human or mouse muscle samples was analysed by RT-PCR focusing on muscle-specific exon 17a expression. To determine the exon composition of only exon 17a-containing isoforms, a sense and antisense primers within the exon were used; the facing primers were designed within exons 2 and 22 sequences, which have been so far described as constitutive sequences (Fig. 6A). A single PCR product was obtained in the upstream region; it contained exons 4–13, 16, and 17. In the downstream region, a major product containing exons 18–22 was obtained. Cloning and sequencing revealed two isoforms, one containing the full-length exon 17a, and one the alternate 17a′ motif. In addition to these major isoforms, a minor product containing exon 17b in association with either 17a or 17a′ was obtained (Fig. 6A).
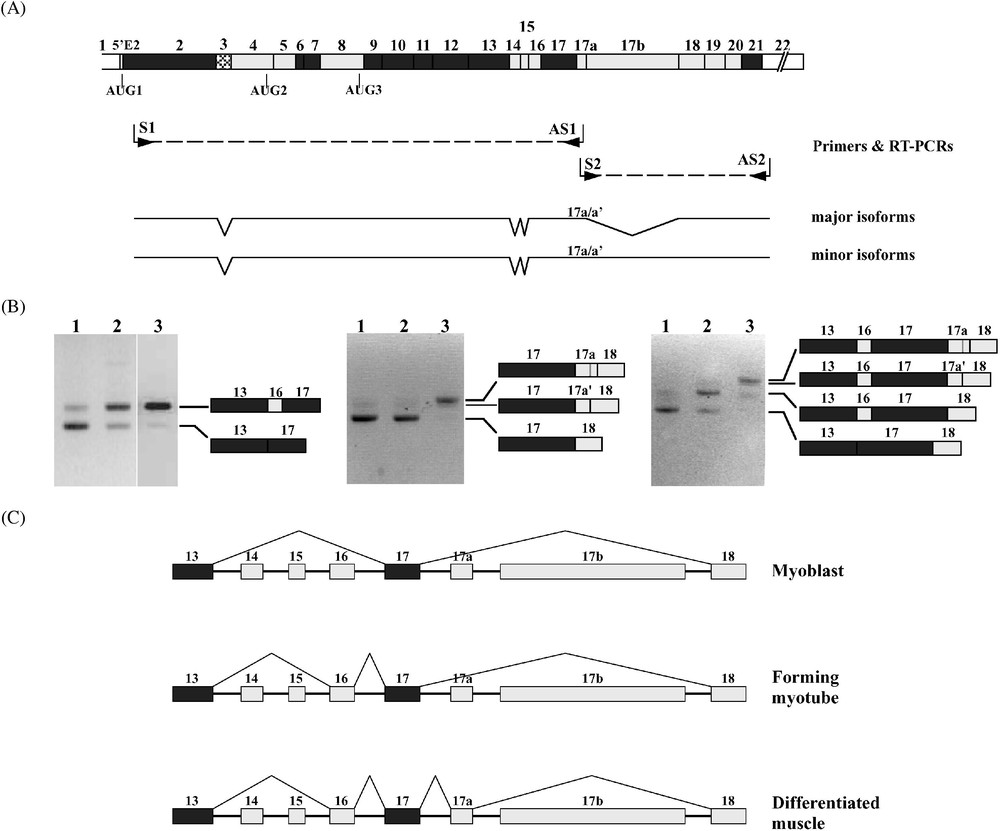
4.1R mRNA alternative splicing in muscle. (A) To select the 4.1R mRNA isoforms bearing the muscle-specific exon 17a, two complementary RT-PCRs were performed using either an antisense (AS1) or a sense (S2) primer within exon 17a, and facing primers (S1 and AS2, respectively) within constitutive exons. The major isoforms lack exon 17b, whereas the minor isoforms contain this exon. However, both species with 17a or 17a′ (17a/17a′) were found in each class of isoforms. (B) 4.1R mRNA from mononucleated myoblasts (samples numbered 1), forming myotubes (samples 2), and mature muscle tissue (samples 3) were amplified by RT-PCR. The PCR products were resolved on 4% NuSieve agarose gels. The different bands were sequenced. Their exon composition is indicated. (C) These experiments led to draw the splicing pathways of the SAB domain-encoding exons at different stages of muscle development.
Five alternatively spliced exons (exons 14, 15, 16, 17a, and 17b) are clustered at the region of 4.1R mRNA encoding the SAB domain (Fig. 6A). We examined the splicing pattern of these sequence motifs in cultured C2C12 myoblasts and during myotube formation, as well as in mature muscle tissue (Fig. 6B). RT-PCR amplification revealed only two major isoforms at exon 13-exon 17 region, one lacking all exons 14, 15, and 16, referred to as isoform , and one isoform , missing exons 14 and 15, but containing exon 16 (Fig. 6B, left panel). The latter is present at a low level in myoblasts; it increases in abundance at the expense of the shorter species to become the major isoform upon myotube fusion and in mature muscle. mRNA analysis of exon 17-exon 18 region of 4.1R transcript revealed a switch from a predominant isoform in myoblasts to a predominant isoform in the differentiated muscle (Fig. 6B, middle panel). However, by contrast with exon 16's early retention, inclusion of exon 17a occurs only in mature differentiated muscle. Additional RT-PCR experiments subsequently confirmed that the appearance of exons 16 and 17a occurs on the same mRNA molecule during myotube formation and in mature muscle (Fig. 6B, right panel), hence confirming our previous data [7], and the data obtained in long RT-PCR experiments (Fig. 6A). RT-PCR using long polymerisation extension step led to amplify the minor isoforms containing exon 17b, in addition to exons 16 and 17a in differentiated muscle samples. These results suggest an asynchronous regulation of 4.1R pre-mRNA alternative splicing of exons at the SAB-encoding domain during muscle differentiation. A similar feature has been reported on 4.1R pre-mRNA, dealing with the early exclusion of 5′E2 motif and the late inclusion of exon 16 during erythroid differentiation [12]. Altogether, these findings allow us to propose an mRNA alternative splicing map of the SAB domain-encoding region of 4.1R mRNA during muscle differentiation (Fig. 6C).
4 Discussion
It is well recognized over the last decade that connection of the cytoskeleton to the basement membrane in skeletal muscle involves two sarcolemma protein complexes: integrin complex and the dystrophin–glycoprotein complex (DGC). The DGC is composed of transmembrane and peripheral membrane proteins [23]. Complete deficiency or structural alteration of dystrophin results in a secondary reduction at the sarcolemma of DGC components such as α-dystroglycan and sarcoglycans [23]. Therefore, the secondary absence of 4.1R in DMD sarcolemma and its colocalization with residual dystrophin in BMD sarcolemma (present work) suggest that 4.1R might be an intrinsic component of a protein complex that associates with dystrophin. Interestingly, the complete absence of erythroid 4.1R exhibit no muscle phenotype in patients with hereditary elliptocytosis [24,25], or in knock-out mice with invalidated 4.1R gene [26]. Similarly, disruption of other genes encoding sarcolemma proteins, such as syntrophin, exhibits no evidence of muscular dystrophy, although a direct binding with dystrophin has been established [27,28].
Muscle fibres are dynamic structures capable of altering their molecular composition and contractile properties in an adaptive manner. Myosin-based classification led to histochemical delineation of type I (slow) and type II (fast) as the major fibre types [29]. Besides myosin isoforms, which are a key marker of fibre-type transitions, a fibre-specific distribution of protein isoforms has been documented in several examples, such as troponin T (TnT), α-actinin and parvalbumin, among others [29]. In the present work, we show that 4.1R(+MSP) isoform, which contains a muscle-specific peptide encoded by an alternate exon, is predominantly expressed in fast myofibre myoplasm. These findings suggest the presence of a coordinate fibre-type-specific program of mRNA splicing regulation.
Early works have documented 4.1R interaction with myosin in red cells [30]. More recently, 4.1R has been shown to interact in muscle with contractile components myosin, α-actin and α-tropomyosin in a highly specific complex [18]. 4.1R is an actin- and spectrin-binding protein. It is therefore not surprising to find this protein both at the sarcolemma and the myoplasm, suggesting a dual function through interactions with actin and spectrin-like (dystrophin) on one side, and through the contractile apparatus on the other side.
Exon 16, first described as an erythroid-specific motif, is in fact predominantly included in muscle 4.1R mRNA isoforms [7]. As a result, the whole SAB domain is expressed in muscle. Interestingly, muscle also expresses a β-spectrin isoform (β1Σ 2, [31]), that shares with the erythroid isoform β1Σ 1 the same N-terminal region [32], which is precisely the binding site for 4.1R in red cells. These findings led to the hypothesis that 4.1R in muscle may also function as a component of a protein complex including β-spectrin. Nevertheless, data gathered from previous works and from the present work argue against this hypothesis: although spectrin distributes together with other membrane skeletal proteins in abnormal sarcolemmal structures, it still associates with the sarcolemma in human and mouse dystrophin-deficient muscle fibres [33–36]. Using an anti-spectrin antibody, we reproduced this observation in human DMD muscle (Fig. 2I–L). On the contrary, 4.1R antibodies labelled the myofibre sarcolemma in normal but not in DMD muscle.
In the present study, we show that the major isoform 4.1R(+MSP) contains the SAB domain encoded by exons 16 and 17, along with the MSP encoded by exon 17a. This isoform is generated through a stepwise-regulated alternative splicing that results in the inclusion of exon 16 in differentiating myotubes, followed by the inclusion of exon 17a in mature muscle cells. However, functional assays tend to suggest that the interaction of 4.1R with myosin involves the 10-kDa SAB domain, mostly through exon 17-encoding peptide [18], and therefore the functional significance of the MSP in 4.1R still remains to be elucidated. In addition to 4.1R isoforms ([7,18], this work), skeletal muscle contains multiple isoforms related to red cell membrane skeletal proteins, such as spectrin [37–40], and ankyrin [41–43]. Most remarkably, spectrin, ankyrin and 4.1R all display a complex, and in some cases fibre-type specific pattern of expression. Further understanding of the expression patterns of protein 4.1R isoforms and of their biochemical capacity to interact with protein complexes will be important for refined understanding of their biological roles in muscle tissue.
Acknowledgments
We are most grateful to Dr M. Garbarz for his generous gift of antibodies to protein 4.1R, to Prof. J.-C. Kaplan for his constant support, and his group for helping with the muscle Western blot technique, and to Prof. A. Morin for kindly providing some muscle biopsies. This work was supported by the ‘Association française contre les myopathies’, the ‘Ligue contre le cancer’, and the ‘Institut national de la santé et de recherche médicale’.


