I joined François’ laboratory in November 1984, coming from the University of Pisa, as part of the exchange program between the Scuola Normale of Pisa and the Ecole Normale Supérieure (ENS). This was after completing my third year of University, which today would be equivalent to a Master 1 degree. At the time, François was leading the Unit of Biochemistry at the Institut Pasteur and was also directing a laboratory at the Collège de France. The Unit of Biochemistry housed several research groups, primarily focused on the biochemistry of muscle, and later, muscle development: the groups of Marc Fiszman, Robert (Bob) Whalen and Margaret Buckingham, who a few years later became independent and founded her own unit in the same department. Initially, the three groups were interested in the biochemistry and expression of contractile proteins, with Bob and Margaret focusing more on myosins and their mRNAs, respectively, and Marc on tropomyosin. I had been accepted to work in Marc’s group.
Marc had set up a nice cellular system whereby differentiation of chicken myoblasts infected by a thermosensitive mutant of the Rous sarcoma virus could be induced by shifting the culture temperature to 42 °C [1]. In Marc’s group the expression of tropomyosin isoforms induced specifically during differentiation was studied biochemically.
But the molecular biology era had started with the isolation of cDNAs that could be used for characterizing the many different mRNAs coding for contractile proteins, for analyzing gene expression and—importantly—for the characterization of genes. Articles had been published by the group of Margaret on the structure of the myosin light chain genes [2, 3].
In Marc’s lab my task was to isolate the gene coding for β-tropomyosin in chicken: we had a cDNA clone provided by David Helfman and in Marc’s group the expression of the isoforms of chicken tropomyosin had been extensively studied during muscle development at the protein level. When we determined the structure of the gene we discovered that it was alternatively spliced, with two couples of exons used in a mutually exclusive manner, exon 6A and 9B in myoblast, 6B and 9A in myotubes. The big question was then how the “right” exons are chosen.
At that time, the understanding of splicing regulation was in its early stages. The discovery that genes were composed of exons and introns had occurred less than a decade earlier [4, 5] and many examples of alternatively spliced genes were emerging, starting in the early and mid 80s [6].
While there was much speculation and research on how the choice of alternatively spliced exons was made, the underlying mechanisms remained largely unknown. This was the challenge we faced. One of my best souvenirs in François’ laboratory revolves around these studies, and reflects the great oversight he provided, even from a somewhat distant eye!
I had analyzed the sequence of the alternatively spliced region around exons 6A and 6B using the available tools of the time, such as early versions of the RNA folding algorithm created by Zuker [7], now called Mfold [8]. I had proposed a model suggesting that exon 6B could not be used in myoblasts because it was sequestered within a secondary structure of the primary transcript [9]. Upon differentiation, an alternative secondary structure would form around the other exon (6A), simultaneously freeing the muscle-specific exon and sequestering the exon used in undifferentiated cells (Figure 1).
RNA secondary structure proposed to sequester exon 6B of the chicken tropomyosin gene to prevent its inclusion in undifferentiated cells (Mb). A scheme of the exonic organization of the gene in this region is shown in the bottom.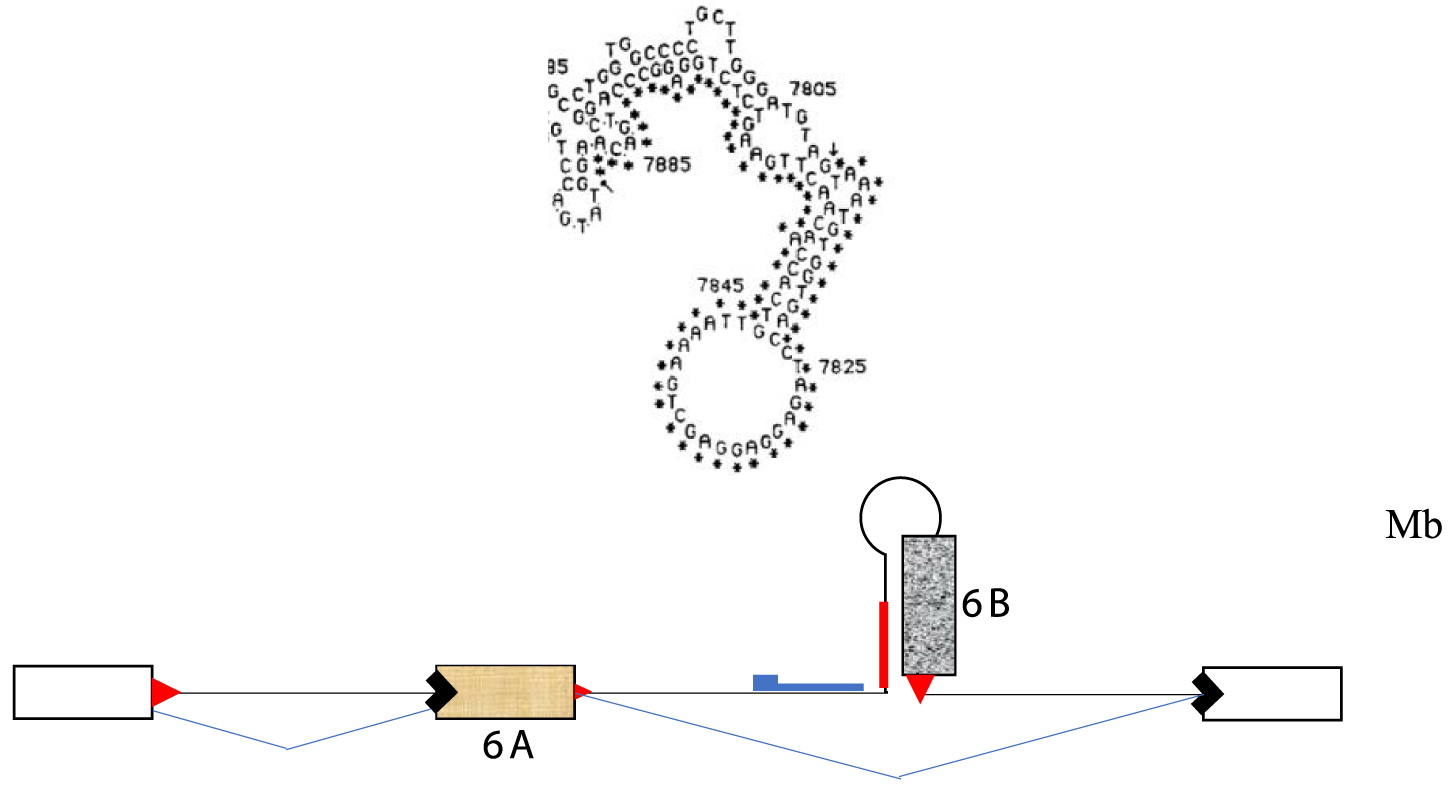
I was very proud of my model, the demonstration of which, in my naiveness, I had taken for granted. Around that time the group of Bernard Dujon had joined the Institute. I remember seeking the opinion of Alain Jacquier, an RNA expert in Bernard’s team and later on an inspirational friend. Alain was very skeptical about the possibility that such a structure would form in the cell, which of course left me in deep despair. I remember walking back to the lab, clutching my now seemingly useless model, and meeting François, who read the disappointment on my face. François was a very busy person, I was not even sure he knew who I was. But, of course, this encounter was a bit like the branch you grab when falling off a cliff, and I started to explain confusedly the reason of my despair. I certainly did not know that François had fallen into the “magic RNA potion” in his youth: “When Monod asked me what I would like to work on, now that I was in his lab … my decision was taken. I would, from then on become an RNA biochemist, an RNA man! Nothing concerning RNA and its role in protein synthesis should be foreign to me.” [10]. François invited me into his office and patiently listened to my story. When I was done, he thought for a couple of minutes, and, of course, agreed with Alain (… falling down the cliff again). He said: “Domenico, ne tombez pas amoureux d’un modèle, tôt ou tard il vous décevra (don’t fall in love with a model, sooner or later it will disappoint you)”—I like to think he was wittily playing with the double meaning of the word “model”. But then he added: “… mais soyez persévérant pour le démontrer (but be perseverant in proving it”.
This was of course the net that saved my fall. Many mutations and compensatory mutations later, the model was partially proven [11], and secondary structures of the primary transcript are now believed to have important roles in alternative splicing [12, 13, 14, 15].
I like to think of this anecdote as nicely summarizing François’ legacy. We should be dreamers, sometimes visionaries, persistent in pursuing demonstrative science, but never in love with models. I am certain that I interpret the thoughts of many colleagues and friends of that period in thanking François for building the great and supportive environment that made possible a very nice moment of science in the Unit of Biochemistry of the Institut Pasteur.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.




 CC-BY 4.0
CC-BY 4.0