1 Introduction
The genes of the classical lac operon of E. coli (lacZ, lacY, and lacA) encode β-galactosidase, lactose permease and thiogalactoside acetyltransferase [1]. Although the catalytic activity of the LacA gene product was initially inferred from the study of thiogalactoside substrates, the subsequent determination of its broader substrate specificity and the more frequent use of the term ‘acetyltransferase’ in recent years has led to the additional designation of LacA as galactoside acetyltransferase (EC 2.3.1.18, GAT). Here, we briefly note some of the early observations that led to the identification and initial characterization of GAT, and focus on more recent work from the field of structural biology that has led to a more complete description of the enzymology of GAT and to the confirmation of its membership in a large superfamily of acyltransferases.
2 Biological role of GAT
Although the roles of the β-galactosidase and lactose permease of the lac operon are well known, there remains some uncertainty about the biological role of GAT. A hypothesized role for GAT as a detoxifying enzyme closely followed the original observations that led to its identification and initial characterization. In the course of investigating the transport properties of methyl-1-thio-β-d-galactoside (TMG), Monod and co-workers reported that about 5% of the extracted intracellular substance was chemically modified [2]. This observation was subsequently extended to additional thiogalactosides, including isopropyl-1-thio-β-d-galactoside (IPTG), with the identification of 6-O-acetyl-IPTG as the chemically altered compound [3]. This acetylated compound did not appear to revert to the original galactoside and did not act as an inducer of the lac operon or as a substrate of lactose permease. This acetyl-CoA dependent acetyltransferase activity was detected only in cell-free extracts obtained from permease-containing strains [4] but did not seem to affect membrane transport [5,6].
Wilson and Kashket reported that E. coli strains accumulated less radiolabeled TMG relative to strains, due to acetylation of TMG, followed by its efflux from the cell and a reduced rate of re-transport of the acetylated compound [5]. The question of whether such a mechanism could confer selective advantage to strains was examined by Andrews and Lin [7], who measured the generation times of E. coli cultures grown on mixtures of metabolizable (lactose or lactulose) and non-metabolizable (TMG or IPTG) substrates. Although IPTG affected the generation times of strains only marginally, the growth rate of strains incapable of acetylating IPTG for discharge into the medium was significantly reduced. These results were extended to mixed culture experiments in which a 15-fold enrichment of vs. cells was observed after 50 generations in the presence of hydrolyzable β-galactosides and IPTG, demonstrating a selective advantage conferred by the genotype under these conditions. Taken together, these experiments led to the hypothesis that the lacA gene serves as a backup device to avoid metabolic congestion, by a mechanism of acetylation, diffusion of the chemically altered compound from the cell, and a diminished rate of retransport. This strategy could allow cells to avoid metabolic predicaments, such as for molecules that are incidentally transported by the broad specificity lactose permease, but which are either non-metabolizable or act as gratuitous inducers. This hypothesis has no significant competition, but has yet to be supported by the identification of any natural high affinity substrate for the enzyme, although it has been argued that high Km values for acceptor substrates could be of adaptive significance if they enabled the cell to use energy to acetylate compounds for discharge only when the levels of the acceptor substrate were high [7].
3 Kinetics and substrate specificity
The partial purification of GAT was first reported in 1962, and permitted a confirmation of its activity against a panel of galactosides as well as the identification of lactose, maltose, galactose and glucose as poor substrates [8]. Subsequent high-level purification of the enzyme led to its crystallization from solutions of ammonium sulfate [9]. The introduction of Ellman's reagent (dithiobis(2-nitrobenzoic acid), DTNB) improved the sensitivity of the assay for GAT by about 100-fold [10], and remains the primary means for the assay of GAT today.
The detailed kinetic mechanism of GAT was first studied by Musso and Zabin and found to be consistent with a sequential ordered bi-bi addition pattern, with acetyl-CoA as the first substrate to associate with the enzyme and CoA as the final product to depart [11]. Galactosides and, to a lesser extent, glucosides (epimers at C-4) were acceptors, but mannosides were not – indicating that the epimeric configuration of the C-2 glycosyl moiety that differentiates these sugars is an important determinant for substrate recognition. These authors also confirmed that the enzyme requires a thioglycoside or a hydrophobic aglycone for activity, in agreement with previous results [8,10]. The measured value for IPTG of 0.77 M based on concentrations of IPTG below those for which substrate inhibition was observed (about 1.5 M) immediately raised suspicion that additional structural features of the biological acceptor for GAT had yet to be identified [11].
The pattern of substrate specificity has more recently been studied by Shaw and co-workers in the course of a kinetic, mutagenic and spectroscopic investigation [12]. This work identified PNPβGal (p-nitrophenyl-β-d-galactopyranoside) as the best substrate, with a value about 10-fold less than IPTG (cf. 63.4 mM vs. 0.77 M). The similar and kcat values for the PNPβGal and phenylβGal substrates suggested that the hydrophobic phenyl group and not the p-nitro moiety of PNPβGal was the primary specificity determinant responsible for the lower Km values of phenyl galactosides relative to IPTG, a molecule bearing an isopropyl aglycone.
4 Structure
4.1 Primary structure
The amino acid sequence of GAT was initially determined by chemical methods [13] and later confirmed in the course of completing the DNA sequence of the lac operon [14]. Based on the sequence of the nodL gene of Rhizobium leguminosarum, Downie detected significant sequence similarity between the amino acid sequence of NodL and the acetyltransferases encoded by the cysE (serine acetyltransferase) and lacA genes of E. coli [15]. NodL acetylates the 6-hydroxyl position of the non-reducing terminal sugar of a variety of lipo-oligosaccharides, chitin fragments and N-acetylglucosamine [16].
Further analysis of these and additional sequences available several years later allowed two groups to identify a previously unrecognized six residue repeated motif in the sequences of these proteins, as well as several other acyltransferases. This repeating sequence theme was termed an ‘isoleucine patch’ [17] or ‘hexapeptide repeat’ [18]. Imperfect copies of this hexapeptide repeat motif, generally described as [LIV]–[GAED]–X–X–[STAV]–X, have since been found as an easily identifiable and characteristic feature of an expanding family of acyltransferases. These hexapeptide acyltransferases catalyze the transfer of acetyl, succinyl or R-3-hydroxy fatty acyl groups from their corresponding thioesters to amino acids, sugars, metabolic intermediates or natural product acceptors bearing free hydroxyl or amine groups. These enzymes participate in the processes of cell wall biosynthesis, amino acid metabolism and detoxification and are represented in all three kingdoms of life, but have yet to be found in animals.
4.2 Overall three-dimensional structure
Although the crystallization of GAT from solutions of ammonium sulfate was first reported by Zabin and co-workers in 1963 [9], the production of crystals suitable for a three-dimensional structure determination (also obtained from solutions of ammonium sulfate) was described 36 years later [19]. These crystals led to an X-ray crystallographic structure determination of GAT in several complexes with substrates and products at resolutions ranging from 3.2 to 2.5 Å [20]. Crystallization of the GAT apoenzyme could not be reproduced to form crystals suitable for crystallographic analysis, nor could back soaking of crystals prepared in the presence of acetyl-CoA be used to yield crystals that were free of cofactor. Hence, the structure of the GAT apoenzyme remains unknown.
The overall structure of GAT is trimeric (Fig. 1) [20]. Its appearance is dominated by a coiled structural domain in the residue range 58–173 of the 203 residue polypeptide that corresponds to its tandem-repeated copies of hexapeptide repeats. This left-handed parallel β-helix (LβH) structural domain [21] is intimately related to the active sites of all of the enzymes belonging to the hexapeptide acyltransferase superfamily of enzymes [20–27]. Although some reports have suggested a weak amino acid sequence similarity between GAT and the classical CATIII chloramphenicol acetyltransferase, their overall conformations are not similar [20,28].
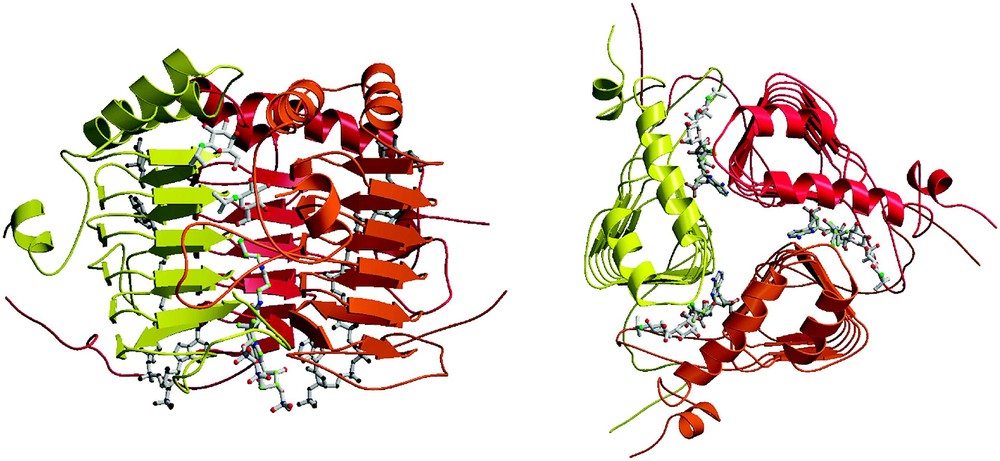
Ribbon diagram of GAT bound to IPTG and CoA. Two IPTG and one CoA molecules are present in each of three active sites of the trimeric enzyme. Left: Viewed perpendicular to the molecular threefold axis. Right: Viewed parallel to the threefold axis, emphasizing the triangular LβH helical domains.
Each six-residue hexapeptide repeat is used to form one side of a triangular coil with a canonical 18 residue length (Fig. 2). Because each coil is composed of an integral number of residues, they are spatially related to one another by simple translations. The overall appearance of the LβH structural domain is that of an equilateral prism, with each flat surface representing a single untwisted parallel β-sheet [21]. The active sites of these trimeric enzymes are invariably located at the junction between two adjacent LβH domains and frequently incorporate one or more polypeptide loops that project from the vertices of their triangular coils. The LβH domain of GAT is composed of approximately 5.3 coils and is interrupted by an extended loop (residues 112–131) (Fig. 2). This loop of 20 residues substitutes for a two residue turn and may have arisen by an evolutionary mechanism by which a single coil of the LβH was expelled from the coiled domain, perhaps by mutation of a residue that violated the hexapeptide repeat sequence rule.
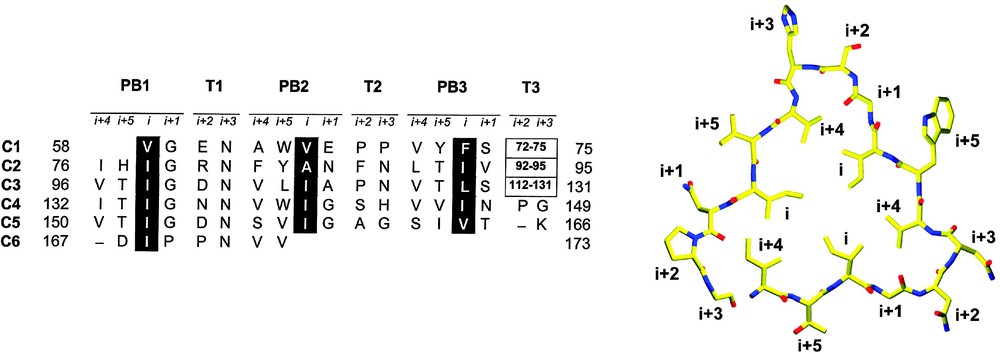
The LβH domain and the hexapeptide repeat sequence motif. Left: Structure-based sequence alignment of the LβH domain of GAT. Residues corresponding to six complete or partial coils (C1 to C6) are depicted as are the residues in this range that lack coiled conformation (boxed), including the extended loop (residues 112–131). Each complete coil is composed of three flat β-sheets (PB1, PB2, PB3) separated by three turns (T1, T2, T3). The residue types repeated within each coil are termed i,i+1,…,i+5 and corresponding to the canonical hexapeptide sequence motif [LIV]–[GAED]–X–X-[STAV]–X, respectively. The most highly conserved residues at the i position are reverse shaded. Right: Coil C4 of the LβH domain with labeled residue types. Residues at the i and i+4 positions project into the lumen of the LβH. Each side of the triangular coil corresponds to a single parallel β-strand.
4.3 Active-site structure and catalytic mechanism
The structure of GAT has been determined in binary complexes with CoA or acetyl-CoA, and in ternary complexes with IPTG/CoA and PNPβGal/ CoA [20]. The GAT trimer contains three apparently independent active sites, each receiving contributions from two subunits, here termed A and B. The conformation of the cofactor resembles a fishhook. Its -phospho ADP moiety is located nearest the C-terminal coils of the LβH and its pantetheinyl arm is directed toward the NH2-terminal coils (Fig. 1). The pantetheinyl arm of the cofactor accepts hydrogen bonds from adjacent coils of the LβH, promoting its extended conformation.
The general location of the IPTG or PNPβGal acceptor substrates is near the CoA thiol or acetyl-CoA thioester group and the galactosyl moieties of these acceptors utilize a conserved pattern of interactions with the enzyme (Fig. 3). The structure of GAT in complex with CoA and IPTG revealed two molecules of IPTG per active site, separated by 8 Å. These molecules are termed proximal or distal, based on their distance to the cofactor thiol. The proximal IPTG forms hydrogen bonds to the enzyme from each of its four hydroxyl groups. The active site of GAT is similar to that of the recently determined structure of E. coli maltose acetyltransferase (MAT) [27]. The interaction of the C-4 hydroxyl group of IPTG with Asp 93B of GAT may confer substrate preference for galactosides over glucosides, since the active site MAT, which preferentially acetylates the 6-hydroxyl group of glucosides, substitutes valine at this position, as does the NodL acetyltransferase capable of acetylating glucose-derived N-acetylglucosamine [16].
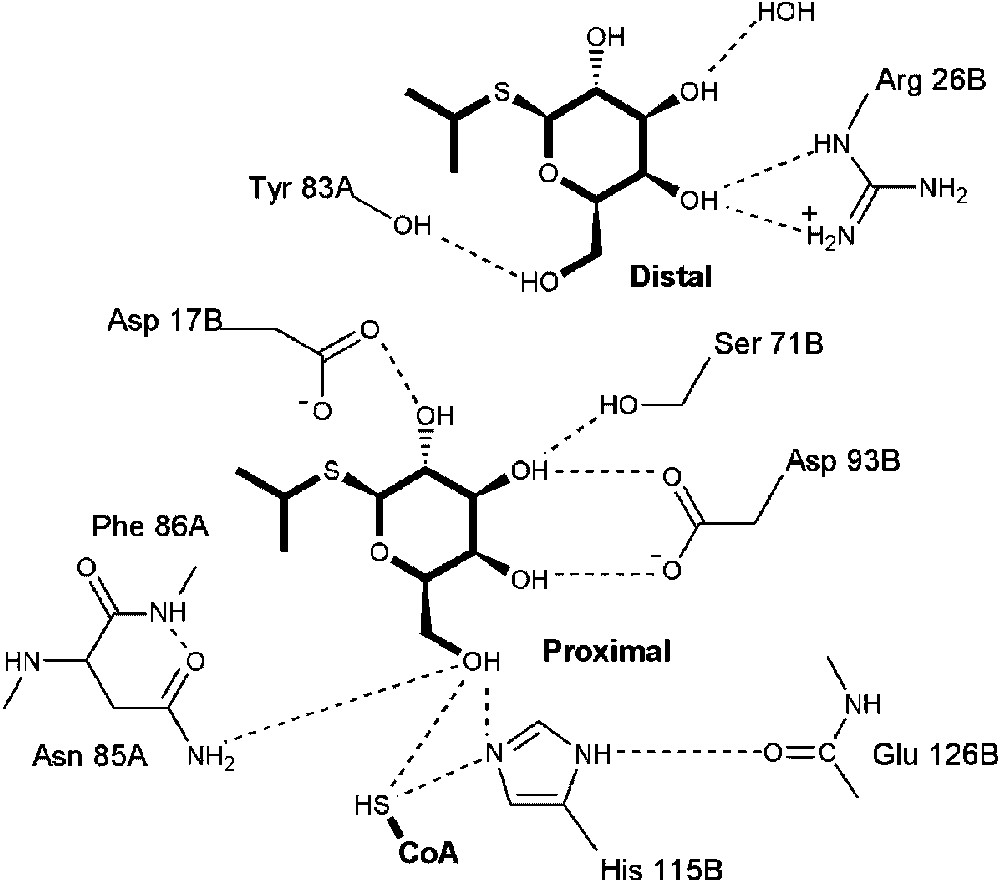
Active-site structure of GAT bound to IPTG and CoA. Hydrophilic interactions are depicted by dotted line segments. The proximal IPTG that participates as the acetyl acceptor is depicted as is the additional distal IPTG molecule.
The distal IPTG is too far from the cofactor to participate as a substrate. Although the concentration of IPTG used to prepare the ternary complex crystals of GAT with IPTG/CoA was high, 156 mM, the existence of the distal IPTG binding site might nonetheless indicate the existence of an extended acceptor binding pocket that could accommodate larger substrates. Whether this distal binding site is also related to the observed phenomenon of kinetic substrate inhibition is unknown.
The hydrophobic binding pocket responsible for binding the aglycone moiety of the acceptor substrate is formed by Tyr 83A, Leu 103A and Met127B (Fig. 4). A biologically important feature of this pocket may be to discriminate against molecules that are not to be acetylated – such as disaccharides and glycosides that bear hydrophilic groups at the same position as the hydrophobic aglycones of IPTG, TMG or PNPβGal. Among the molecules that are poor substrates of GAT are lactose (4-β-d-galactosyl-d-glucose) and two natural inducers of the lac operon-allolactose (6-β-d-galactosyl-d-glucose) and the plant product, 2-β-d-galactosyl-glycerol, all of which bear hydrophilic substituents at this position.
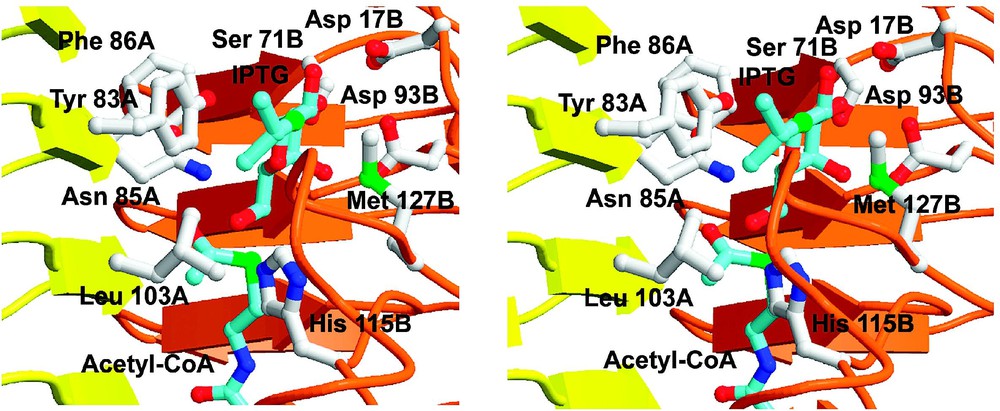
Superposition of GAT in complex with acetyl-CoA and IPTG (blue bonds). The residues referred to in the text are labeled. Subunits A (yellow) and B (orange) are depicted.
GAT catalyzes the acetyl-CoA dependent acetylation of the 6-O-methyl position of a variety of pyranosides. The 6-hydroxyl group of the proximal IPTG in the IPTG/CoA complex interacts with His 115B from the extended loop, Asn 85A, and the thiol group of CoA. His 115 had been previously implicated in the iodoacetamide inactivation of the enzyme and its replacement by alanine reduced kcat by 1800-fold [12]. His 115 and Asn 85 are also conserved in the sequences of MAT and NodL. In GAT, this histidine donates a hydrogen bond to the peptide carbonyl oxygen of Glu 126B, which identifies its imidazole ND1 nitrogen as protonated and presumably increases the basicity at NE2 (Figs. 4, 5). The distance between the phenyl ring of PNPβGal or the isopropyl group of IPTG to His 115 is only 4 Å, and so it remains possible that some molecules may bind at or near the active site of the enzyme, but do so in such a manner as to alter the orientation of this histidine residue to prevent efficient catalysis.
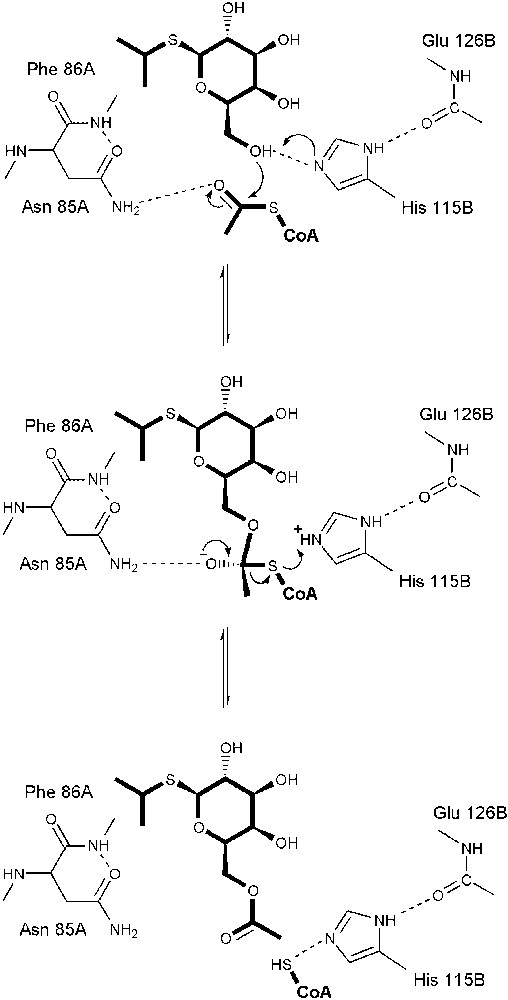
The proposed SN2 reaction mechanism of GAT derived from kinetic and structural studies. His 115B functions to abstract a proton and perhaps to protonate the resulting thiolate. A tetrahedral intermediate is formed by the attack of the IPTG 6-hydroxyl group on the thioester carbonyl carbon acetyl-CoA and is stabilized by the carboxamide group of Asn 85A.
The kinetic mechanism of GAT is best described as sequential ordered bi-bi [11,12], consistent with a chemical mechanism whereby the thioester group of acetyl-CoA is attacked directly by the 6-hydroxyl group of the substrate. This is supported by the crystal structure of GAT in complexes with acetyl-CoA and IPTG/CoA (Fig. 4). A superposition of these structures places the 6-hydroxyl group of the acceptor within 1.7 Å of the acetyl-CoA thioester carbonyl carbon atom, indicating that this hydroxyl group may indeed be close enough for direct attack in an SN2 ternary complex mechanism. Taken together with kinetic data and the structure of GAT in complex with acetyl-CoA, a chemical mechanism of action can be proposed in which His 115B acts to extract the 6-hydroxyl proton from the acceptor prior to or concomitant with attack of this group on the thioester. The observed interaction of Asn 85A with the acetyl-CoA thioester oxygen may indicate that this side chain plays a role in stabilizing the oxyanion intermediate. His 115B may also function to donate a proton from the NE2 position to the resulting CoA thiolate concomitant with or subsequent to the collapse of the tetrahedral intermediate.
4.4 Relationships to other hexapeptide acyltransferases
It is now well-established that GAT is a member of the hexapeptide acyltransferase superfamily of enzymes bearing tandem-repeated copies of a six-residue periodicity theme. These hexapeptide repeats encode folding of an unusual coiled LβH domain which is intimately involved in the construction of the active sites of these enzymes. However, the active sites of these enzymes also accept contributions from portions of the polypeptide chain external to the LβH and so the detailed arrangements of active site residues differ significantly within this superfamily [20–27]. Of the hexapeptide acetyltransferases of known three-dimensional structure, GAT is most similar to MAT, with 42% amino acid sequence identity and an rms deviation for 169 selected Cα coordinates of only 1.0 Å [27]. Although MAT does not belong to an obvious operon, its proposed role is also as a detoxification agent, with preference for non-metabolizable glucosides bearing a hydrophobic aglycone [27,29]. The best known substrate for MAT is isopropyl-1-thio-β-d-glucoside (IPTGlu), the C-4 epimer of IPTG, with a Km value of 17.2 mM [27]. In addition, the GAT, MAT and NodL members of the hexapeptide acyltransferase superfamily all acetylate the 6-hydroxyl group of hexose sugars and conserve His 115 and Asn 85 at their active sites.
5 Conclusions
Recent structural characterization of GAT has revealed its overall three-dimensional structure, substrate binding modes and catalytic mechanism. Consideration of its amino acid sequence and the crystal structures of a variety of acyltransferases has confirmed its placement in the hexapeptide acyltransferase superfamily of enzymes. The similarities of its sequence, substrate specificity and active site structure to MAT and NodL define a subgroup of the hexapeptide acyltransferase superfamily whose members transfer acetyl groups from acetyl-CoA to the 6-hydroxyl group of hexose sugars, probably by a common catalytic mechanism. A current gap in knowledge exists for GAT as it is not known which substrates the enzyme encounters in vivo. Until this gap is filled, it is likely that some doubt will remain concerning the biological role of this member of the lac operon.
Acknowledgments
We thank Dr. Xing-Guo Wang for assistance and helpful discussions. This work was supported by the National Institutes of Health Grant AI-42154.


