1 Introduction
The initiation of replication of chromosomal DNA is coordinated with cell division. It has been proposed that DNA replication in bacterial cells is initiated on membranes and that the activities of replication proteins are regulated by membrane components [1].
Indeed, initiation of replication of the Escherichia coli chromosome is precisely regulated in the cell cycles [2]. The SeqA protein seems to be one of the key proteins in the control of this process [3–5]. It is an inhibitor of the onset of E. coli chromosome replication in vivo [6,7] and, at high concentrations, of the replication initiator protein, DnaA, in vitro, but it may stimulate replication at low DnaA concentrations in vitro [5]. It affects DNA topology and inhibits open complex formation at the replication origin [8,9]. It was demonstrated in vivo that SeqA limits DnaA activity in replication from the chromosomal origin oriC [4]. Moreover, SeqA protein is essential for sequestration, which affects oriC in the newly replicated hemimethylated state [3,6].
Until methylated by Dam methyltransferase [10,11], the newly replicated GATC sequences exist in a hemimethylated state that in the parental strand is methylated and in the nascent strand is not. Methylation of GATC sites by Dam methylase is implicated in regulation of E. coli chromosomal replication. In fact, the seqA gene was found to be responsible for the sequestration of hemimethylated oriC [6].
The coordination of the synchronization of the replication initiation, the activation of the DnaA protein at oriC and the cellular cycle suggested the existence of a very narrow interaction between the bacterial membrane [12], the protein Dam methyltransferase [13] and the SeqA protein [14].
Acidic phospholipids, such as cardiolipin (CL) and phosphatidylglycerol (PG), decrease the affinity of adenine nucleotide for DNA protein [15,16]. Thus, it has been proposed that phospholipids regulate the activity of DnaA protein in cells and in vitro [15,17].
It has been reported recently that the transcription of genes implicated in lipid and phosphatidic acid biosynthesis was increased in dam mutant [18]. Therefore, Dam methyltransferase appears to be involved in the transcriptional regulation of the metabolism of fatty acids and phospholipids.
In addition, it has also been demonstrated that the seqA mutation can overcome the incompatibility phenotype observed between the chromosomal oriC and minichromosomal oriC copies in the dam mutant strain [19]. The mutation in the seqA gene allows efficient transformation of fully methylated minichromosomes into dam mutant cells [3,4] and the accumulation of seqA and dam double mutation can re-establish partially the replication asynchrony observed at every seqA and dam mutant.
We can suggest a possible interaction between the activities of SeqA and Dam methyltransferase proteins and membrane phospholipids composition.
We speculated that examination of fatty acid composition and phospholipids fractions in dam and/or seqA mutants would provide useful information for understanding the influence of mutations in dam and seqA genes on the fatty acid and phospholipids metabolisms in E. coli.
2 Experimental procedures
2.1 Bacterial strains used
The E. coli strains and their genotypes used in this study are listed in Table 1. Pre-incubated suspensions of E. coli cells were diluted 100-fold with Luria-Bertani (LB) medium and cultured at 30 °C. Exponentially growing E. coli cells in LB medium were harvested by centrifugation when the optical density at 600 nm reached 0.5.
Bacterial strains used in this work
| Strains | Genotypes | References |
| C600 | LacY1, leuB6, supE44, thr-1, tonA21 | [20] |
| C600 dam13 | LacY1, leuB6, supE44, thr-1, tonA21, dam13::Tn9 | [10] |
| C600 ΔseqA | LacY1, leuB6, supE44, thr-1, tonA21, ΔseqA | [3] |
| C600 dam−seqA− | LacY1, leuB6, supE44, thr-1, tonA21, dam13::Tn9, seqA::Tn3 | [4] |
2.2 Analysis of fatty acids in total lipids and phospholipids
The different bacterial strains were cultured at 30 °C and cells were harvested from 1000-ml cultures in exponential phase by centrifugation (4000 g, 10 min, 4 °C), washed with 1% NaCl and total lipids were extracted by the method of Bligh and Dyer [21].
Fatty acids were converted to their methyl esters (FAMEs) according to the method described by Cecchi and collaborators [22]. An aliquot of the solution was evaporated; then 2 ml of hexane, a known quantity of heneicosanoic acid methyl ester [C21:0] as an internal standard and 0.5 ml of sodium methylate (1%) were added. After stirring over 1 min and standing for 2 min, the mixture was neutralized by 0.2 ml of H2SO4 (1 N); then, the methyl esters were washed with 1.5 ml of distilled water. The upper phase solvent containing the FAMEs was removed under vacuum.
Fatty acids relative composition was subsequently determined as % of the total fatty acids using a HP 6890 chromatograph equipped with a flame ionisation detector (FID) and an electronic pressure control (EPC) injector. A polyethylene glycol fused silica capillary column (Innowax, film thickness) purchased from Agilent (Wilmington, Delaware, USA) was used.
The column was operated at 150 °C for 1 min, and the temperature was raised by 15 °C min−1 to 200 °C and finally held at 242 °C for 2 °C min−1. N2 was used as the carrier gas at a flow rate of 1.6 ml min−1 and the split ratio used was 60:1.
Total lipids were placed as thin stripes on thin-layer silica gel plates and resolved by ascending chromatography using chloroform–acetone–methanol–acetic acid-water (50:20:10:10:5, vol:vol:vol:vol:vol) [23]. Standards of various commercial phospholipids (Sigma) were run simultaneously. The phospholipids were visualized with I2 vapours and their bands were marked and scrapped off the plates; the phospholipids were eluted and their transmethylated fatty acids analysed as described above.
3 Results
3.1 Phospholipid composition of the seqA mutant membrane
In this study, we analysed phospholipid compositions of the bacterial membrane with the aim of correlating the membrane structure variation in phospholipids and in fatty acids, already suspected by Wegrzyn and collaborators [24], with seqA gene mutation.
Indeed, the phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) and cardiolipin (CL) proportions were affected by the seqA mutation while comparing them with the wild-type strain that have been incubated at 30 °C.
The major phospholipids present in E. coli wild-type strain membrane were phosphatidylethanolamine (PE), accounting for about 78% of total phospholipids, followed by phosphatidylglycerol (PG) and cardiolipin (CL) (9% and 3% of total phospholipids, respectively) (Fig. 1).
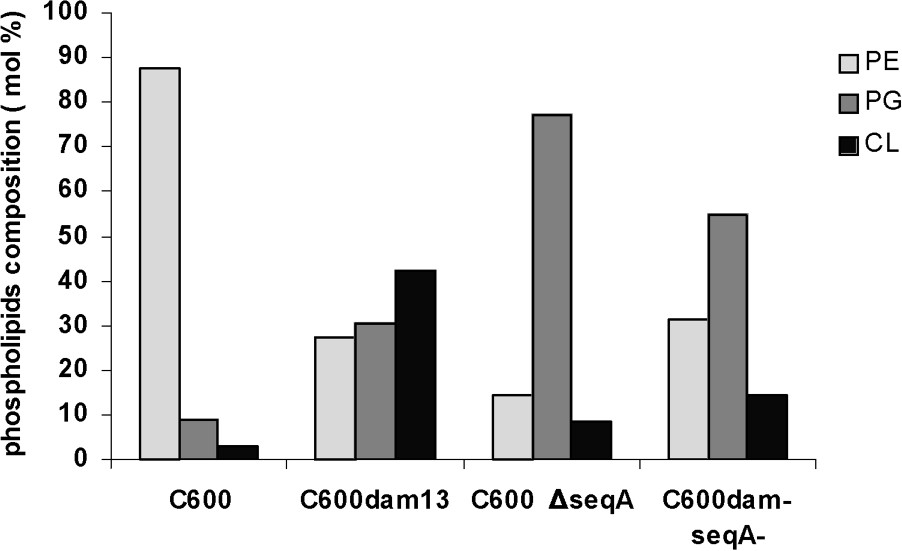
Comparative analysis of phospholipids level in four isogenic strains for E. coli. Percentage of phospholipids analysis from C600 (wild type), C600 dam13 (dam13::Tn9), C600 ΔseqA and C600 dam−seqA− (dam13::Tn9, seqA::Tn3). Exponentially growing C600 (wild type), C600 dam13, C600 ΔseqA and C600 dam−seqA− were incubated at 30 °C, centrifuged when the optical density at 600 nm reached 0.5. After extraction of the total phospholipids, individual phospholipids were separated by solid-phase extraction. Their contents were then calculated from the fatty acid contents measured by the capillary GC method, as described in the § Experimental procedures. Average values of duplicates are given, and the deviation was less than 5% of each value.
In the seqA mutant, the acidic phospholipid fraction (PG and CL) becomes majority of total phospholipids with 85.7%, distributes in 77% of PG and 8.7% of CL (Fig. 1).
This result corroborates the cross-feedback model proposed by Shibuya and colleagues [25].
3.2 Fatty acid compositions of the seqA mutant phospholipids
In the wild-type strain (C600) palmitic acid (16:0), palmitoleic acid (16:1), and oleic acid (18:1) were the main constituents, representing 90.1% of the total fatty acids (Table 2).
Fatty acid composition in total lipid and different phospholipid classes in the dam mutant, seqA mutant and in the dam seqA double mutant
| C14:0 | C16:0 | C16:1 | C17:0 | C18:0 | C18:1 | C18:2 | C18:3 | |
| C600 (total) | 1.2±0.21 | 35.7±0.52 | 18.9±1.00 | 0.9±0.15 | 5.5±0.91 | 37.6±0.74 | 0.2±0.05 | 0.2±0.05 |
| C600 (PE) | 7.0±0.35 | 28.3±0.85 | 9.1±0.48 | 0.8±0.09 | 1.9±0.03 | 46.3±0.43 | 6.5±0.53 | 0.0 |
| C600 (PG) | 0.0 | 41.6±0.64 | 11.9±1.02 | 3.7±0.07 | 3.6±0.25 | 29.8±0.41 | 4.7±0.29 | 4.7±0.33 |
| C600 (CL) | 16.0±0.65 | 1.3±0.05 | 21.9±0.70 | 8.9±0.10 | 8.7±0.27 | 26.1±0.56 | 0.3±0.10 | 16.9±0.36 |
| C600 dam13 (total) | 2.2±0.09 | 32.1±0.34 | 23.6±0.53 | 2.2±0.31 | 2.0±0.22 | 35.5±1.35 | 0.5±0.11 | 1.9±0.14 |
| C600 dam13 (PE) | 3.4±0.10 | 43.0±0.42 | 19.9±0.32 | 3.1±0.15 | 1.9±0.23 | 25.1±0.49 | 2.2±0.13 | 1.4±0.03 |
| C600 dam13 (PG) | 2.4±0.10 | 25.5±0.45 | 13.3±0.12 | 5.4±0.03 | 1.3±0.15 | 40.2±0.70 | 9.3±0.26 | 2.6±0.09 |
| C600 dam13 (CL) | 0.0 | 23.6±0.46 | 9.2±0.26 | 10.0±0.15 | 4.1±0.22 | 21.9±0.26 | 25.4±0.87 | 5.9±0.15 |
| C600 ΔseqA (total) | 0.6±0.06 | 31.7±0.44 | 19.6±0.32 | 2.0±0.03 | 3.8±0.22 | 42.0±0.38 | 0.2±0.04 | 0.3±0.01 |
| C600ΔseqA (PE) | 1.5±0.06 | 20.7±0.27 | 7.3±0.21 | 3.8±0.12 | 6.9±0.13 | 32.3±0.46 | 21.3±0.21 | 6.3±0.23 |
| C600ΔseqA (PG) | 0.9±0.06 | 43.3±0.47 | 13.3±0.10 | 1.7±0.06 | 1.9±0.14 | 36.7±0.35 | 1.1±0.01 | 1.1±0.07 |
| C600ΔseqA (CL) | 5.9±0.20 | 43.0±0.73 | 3.2±0.12 | 1.5±0.02 | 4.5±0.10 | 36.0±0.36 | 5.9±0.41 | 0.0 |
| C600 dam−seqA− (total) | 0.7±0.11 | 39.3±0.88 | 14.4±0.36 | 6.7±0.15 | 6.0±0.14 | 32.4±0.36 | 0.5±0.03 | 0.1±0.02 |
| C600 dam−seqA− (PE) | 0.0 | 10.1±0.11 | 3.5±0.37 | 12.8±0.27 | 5.0±0.12 | 43.8±0.33 | 24.8±0.19 | 0.0 |
| C600 dam−seqA− (PG) | 2.9±0.04 | 6.1±0.10 | 2.5±0.17 | 1.4±0.19 | 10.3±0.16 | 51.5±0.53 | 23.3±0.68 | 2.0±0.05 |
| C600 dam−seqA− (CL) | 10.2±0.87 | 24.7±0.52 | 23.4±0.32 | 0.0 | 11.0±0.15 | 13.0±0.25 | 9.8±0.77 | 7.9±0.40 |
In the seqA mutant , the accumulation of the PG is accompanied with a weak variation of the Ratio of unsaturated to saturated fatty acids (Fig. 2). Indeed, the PG relative proportion of saturated C16:0 acid and unsaturated C16:1 and C18:1 acids appeared to be unaffected by the seqA mutation (Table 2).

The ratio of acyl chain-unsaturated fatty acids in different phospholipids classes in the dam mutant, seqA mutant and in the dam seqA double mutant. The ratio of acyl-chain-unsaturated to acyl-chain-saturated fatty acids (mol mol−1) was calculated using the data in Table 2.
Compared to the isogenic wild-type strain, the seqA mutant increases slightly the membrane CL proportion and decreases its level of acyl chain unsaturation of fatty acids (Table 2). This is due to the accumulation of the saturated C16:0 acid and to the reduction of the unsaturated C16:1 acid in the membrane CL proportion.
3.3 Effect of the dam mutation on phospholipids composition
Compared to the isogenic wild-type strain, the PE and PG and CL proportions were strongly affected by the dam mutation. In fact, the acidic phospholipids fraction remains majority with a high increase of the CL fraction (42.2%) of total phospholipids (Fig. 1).
We analysed the fatty acids composition of the dam mutant cardiolipin (Table 2). We notice that the level of acyl chain unsaturation of fatty acids in the CL phospholipid decreases slightly in relation to the wild-type strain. This diminution reflects a reduction of the unsaturated C16:1 and C18:1 fatty acids to the profile of the saturated C16:0 fatty acid (Table 2). We can also detect an accumulation of the unsaturated linoleic acid (C18:2) (Table 2).
However, the PG fraction of the dam mutant membrane shows a meaningful increase of the level of acyl chain unsaturation of fatty acids (Fig. 2). This is due to the reduction of the saturated C16:0 fatty acid and to the slight increase of the unsaturated C18:1 fatty acid (Table 2).
3.4 Phospholipids and fatty acids compositions of the dam seqA double mutant
To evaluate the combined effect of the two mutations in the seqA and dam genes on the bacterial membrane integrity, we compared the phospholipids composition of the double mutant with the isogenic wild-type strain (Fig. 1).
The acidic membrane phospholipid proportion (PG and CL) is in the majority with respect to the zwitterionic phospholipid, PE, in the dam seqA double mutant with 68.8% of total phospholipids distributes in 54.6% of PG and 14.2% of CL (Fig. 1). We can also note that in the dam seqA double mutant, PG and CL acidic phospholipids have intermediate proportions in relation to the seqA and dam simple mutants (Fig. 1).
In the dam seqA double mutant, we observe an accumulation of acyl-chain-unsaturated fatty acids in the PG and PE fractions. Indeed, the PE fraction shows a reduction of C16:0, whereas the PG fraction accumulates about two-fold quantity of unsaturated C18:1 fatty acid in relation to the isogenic wild cells (Table 2).
4 Discussion
Interactions of E. coli Dam and SeqA proteins with cellular membranes have been reported previously [6,12,18,24,26]. However, although regulation of the activities of these proteins by membranes or their components was reported [18] or suggested [6,24], little is known about the influence of Dam methyltransferase and SeqA on the composition of cell membranes. In this report, we have shown that zwitterionic (PE) and acidic phospholipids (PG and CL) proportions as well as their acyl chain unsaturation of fatty acids levels were affected by different mutations in the seqA and dam genes in relation to wild-type isogenic bacteria membrane.
We observed that the dam mutant membrane accumulates a high fraction of the acidic phospholipid cardiolipin (42.2% of the total membrane phospholipid) containing saturated fatty acids. Thus, we might assume that the expression of genes coding for components of the membranes or for proteins involved in synthesis of such components might be impaired in the dam mutant.
This hypothesis is reinforced by the fact that Dam-mediated methylation is known to be responsible for regulating metabolism, replication and mismatch repair in E. coli [11,18]. Since the Dam methyltransferase protein has also been proposed to form and/or maintain chromosome structure [27], it is likely that this protein could affect the expression of many genes, not only by direct activation or repression of particular promoters via interaction with their GATC sequences, but also by changing DNA topology.
Taken together with the earlier observation that dam mutant initiates chromosome replication asynchronously [26] and that this asynchrony mainly results from an inability to inactivate newly replicated origins that have been sequestrated in the membrane [12], we proposed that the initiation of the replication might be controlled by the Dam methyltransferase directly by affecting the methylation status of GATC sequences of oriC and indirectly by regulating the DnaA activation via the modification of the acidic phospholipids composition and their acyl chain saturation of fatty acids of the membrane.
Like the Dam methyltransferase, it has been demonstrated that SeqA also affects DNA topology and inhibits open complex formation at the replication origin [8]. Indeed, Weito and collaborators [28] have shown that the seqA mutation increases negative superhelicity of chromosomal and plasmid DNA and affects profoundly the transcription of various genes [24,29–31]. In addition, SeqA is involved in translocating oriC into the membrane after initiation of DNA replication [3].
So, Dam and SeqA proteins participate in the movement of oriC into the membrane and may interact with the membrane phospholipids. To examine this hypothesis, we have extracted and analysed the phospholipids composition of the seqA mutant membrane as well as its level of unsaturation of fatty acids.
In this report, we have shown that in the absence of the SeqA protein, the bacteria membrane accumulates a high fraction of acidic phospholipids (85.7% of the total phospholipids with 77% the phosphatidylglycerol).
So, we suggest that in addition to its direct role in the sequestration of oriC on the membrane, SeqA can interact with the lipid metabolism and regulate the acidic phospholipids synthesis.
In the dam seqA double mutant, we observe an accumulation of acyl-chain-unsaturated fatty acids in the PG and PE fractions. However, Makise and collaborators [17] have reported that unsaturated fatty acids increase membrane fluidity that may be important for the control of the DnaA binding to oriC in vitro and can be related with the partial re-establishment of the replication asynchrony observed at every seqA and dam mutant [19].
This suggests that Dam methyltransferase and SeqA proteins might play direct roles in fatty acids' and phospholipids' metabolisms and that impairment of functions of both proteins results in less dramatic alterations in these metabolisms.
In conclusion, the data presented in this work indicate that the fatty acids and phospholipids metabolisms are significantly influenced in E. coli dam and/or seqA mutants and it is likely that several different molecular mechanisms are involved in the regulation of the composition of cellular membranes by these proteins.
Acknowledgements
We are much grateful to Dr M. Kohiyama for providing bacterial strains, but for which this work would not have been made possible.


