1 Introduction
Whilst the end of the 19th century and the beginning of the 20th century coincided in microbiology with the recognition of the microbial role in disease and the existence of the immune system, the new millennium will be associated with the availability of the complete bacterial genomes and the recognition that outside the laboratory flask bacteria grow attached to surfaces and embedded in a self-produced extracellular matrix, referred to as biofilms (Fig. 1). Although the synthesis of the matrix is energy consuming, the life in the community protects bacteria against environmental insults, the immune system or the action of biocides. Since most of all the bacterial species are able to produce biofilms, it seems reasonable that unrelated bacteria make use of common elements for biofilm development. In the last ten years, many groups have dedicated great efforts to identify factors involved in biofilm development. As a consequence, it has been discovered that different unrelated bacteria produce: (i) the same exopolysaccharides (cellulose, poly-β-1,6-N-acetylglucosamine) to build the matrix, (ii) the same secondary messenger, c-di-GMP, to regulate the production of biofilm matrix exopolysacharides, and (iii) a group of surface proteins that exhibit homology with Bap of Staphylococcus aureus (Biofilm-associated protein) to promote adhesion to biotic and/or abiotic surfaces (listed in Table 1).
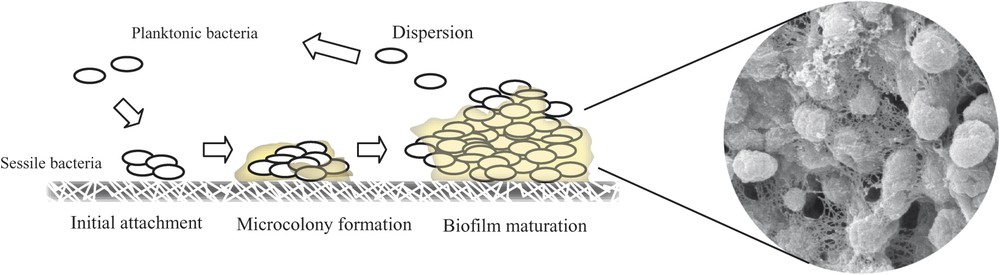
Schematic figure showing biofilm formation process. Visualization of the biofilm formed by Salmonella through Scanning Electron Microscopy.
Biofilm-associated proteins
| Protein | Microorganism | Homology with Bap | Function | Reference |
| Bap | Staphylococcus aureus | – | Primary attachment and biofilm formation on abiotic surfaces | [31,36] |
| Involved in persistent infections | ||||
| Mus 20 | Pseudomonas putida | 8e–136 29% of identity | Initial seed colonization | [25] |
| BapA | Salmonella enteritidis | 3e–114 28% of identity | Biofilm formation | [30] |
| Host colonization | ||||
| Bap | Burkholderia cepacia | 8e–135 33% of identity | Late stages of biofilm formation | [26] |
| Espfm | Enterococcus faecium | 1e–125 27% of identity | [28] | |
| Adhesion to eukaryotic cells | ||||
| Esp | Enterococcus faecalis | 2e–118 26% of identity | Primary attachment and biofilm formation on abiotic surfaces | [24,25] |
| Colonization and persistence in the urinary tract | ||||
| LapA | Pseudomonas fluorescens | 8e–59 25% of identity | Progress to the irreversible attachment stage of biofilm development | [27] |
| Attachment to quartz sand | ||||
| YeeJ | Escherichia coli | 5e–18 21% of identity | Adhesion | [34] |
| VP1443 | Vibrio parahaemolyticus | 3e–05 20% of identity | Biofilm maturation | [28] |
In this review, we will focus on these surface proteins and describe three representative members that mediate biofilm formation in three unrelated bacterial species.
2 Bap in Staphylococcus aureus
The first member of the BAP family was described during a previous investigation carried out by our group focused on the development of the Staphylococcus aureus biofilm. This study, based on a standard screening of transposon mutants unable to adhere to a polystyrene surface, identified a novel 6831pb open reading frame involved in biofilm development [1]. This gene is carried in a putative composite transposon inserted in the SapIbov2 pathogenicity island [2] and encodes for a 2276-amino acids surface protein, named Bap (Biofilm-associated protein). The gene bap has also been detected in isolates of several coagulase-negative staphylococcal species, like S. epidermidis, S. chromogenes, S. xylosus, S. simulans, and S. hycus [3]. However, up today, none of the human S. aureus isolates tested harbour the bap gene, suggesting that ruminant and human staphylococcal strains differ in their host-specific pathogenic strategies.
3 Bap structure
Bap shows structural characteristics typical from cell-wall-anchored proteins present in gram-positive bacteria. The N-terminus domain contains a signal sequence for extracellular secretion (first 44 aa), followed by two separated repeats of 32 aa (aa 45 to 360), which were designated as region A. The remaining amino terminal sequence up to amino acid 818 was designated as region B. According to the Coiled-Coil prediction program, a dimerization domain can be distinguished both in region A (aa 227 to 309) and region B (aa 528 to 548) [4]. This fact suggests that two Bap molecules, belonging to the same or different bacteria, could undergo dimerization and thus promote biofilm formation. However, there is no experimental data to support this hypothesis. The most distinctive structural feature of this protein is the presence of a core region consisting on 13 successive repeats of 86 amino acids, identical even at the nucleotide level [1]. These repeats are predicted to fold into a seven-strand-β-sandwich and likely belong to the HYR module, a new immunoglobulin-like fold involved in cellular adhesion [5]. Although the function of this region has not been discerned, it is hypothesized that it could have a structural role, maintaining the proper protein conformation on the cell surface [1]. It has been demonstrated that the identity existing between the repeats triggers recombination events between them, leading to the production of proteins with different lengths throughout the course of a staphylococcal infection. Curiously, the decreased number of repeats does not affect the protein functionality, even when only one repeat constitutes the core region [6]. The C terminus region comprises three short repeats of 18 aa, designated as region D, and a cell-wall-anchoring region consisting of an LPXTG motif and a series of positively charged residues (Fig. 2).
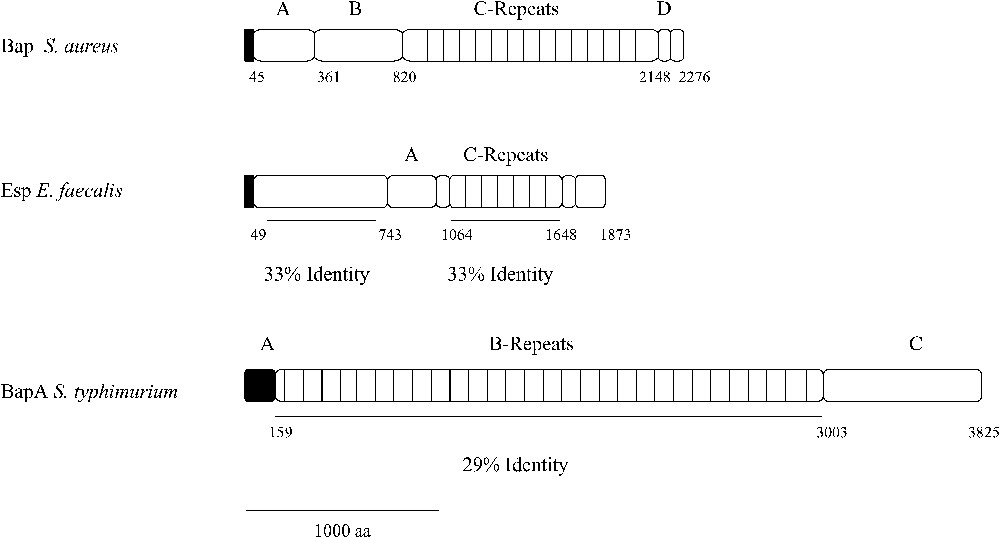
Structural analysis of Esp and BapA and identity to Bap from S. aureus.
It is worth standing out the presence of four potential EF calcium binding motifs within the Bap sequence. Three of them are located N terminal to the C repeat region, whereas the fourth is located at the C terminus close to the LPXTG motif [7]. The role of this calcium binding sites in Bap functionality and biofilm formation will be discussed later.
4 Role of Bap in the biofilm formation process of bovine Staphyloccus aureus
In vitro experiments have shown that Bap is not only involved in the primary attachment step, but also, together with the polysaccharide PIA/PNAG, in cell-to-cell aggregation and thus biofilm maturation. Reinforcing the existence of cooperation between PIA/PNAG and Bap during the second step of biofilm formation, Cucarella and colleagues reported evidence of polysaccharide increased production in Bap overproducing strains and decreased levels of PIA/PNAG in strains whose bap gene had been disrupted [1]. In view of the foregoing, it was supposed that biofilm deficiency of the bap mutant strain was due to an insufficient PIA/PNAG polysaccharide level, but, intriguingly, strains harbouring the gene bap were able to produce biofilm and display a positive phenotype on Congo red agar, even if the ica operon, responsible for PIA/PNAG synthesis, was disrupted [6]. Thus, there is every sign that the expression of the Bap protein is sufficient to promote a PIA/PNAG-independent biofilm development on abiotic surfaces.
5 Role of Bap in pathogenesis
Since its biofilm-forming capacity is widely considered as a major virulence factor of S. aureus, multiple studies have been performed in order to understand the contribution of genes involved in biofilm development to the pathogenic process. In a first approach to evaluate the role of Bap in virulence, a murine catheter-induced infection model was used [1]. The results showed that although the bap mutant strain was more prevalent during the initial stages of infection, later on, the bap positive strain led to biofilm formation and provoked a persistent infection [1]. Following experiments have revealed that strains harbouring bap showed a lower capacity of (i) adhesion to immobilized fibrinogen or fibronectin, (ii) adhesion to mammary gland tissue ex vivo, (iii) internalization by epithelial cells, and (iv) infection of the mammary glands of ewes. Altogether, these results suggest that Bap hinders interaction between bacterial MSCRAMMs receptors and the host proteins, favouring the establishment of chronic infections [8]. Taking into account the fact that bap is replacing the TSST-1 toxin gene (tst) present on SaPIbov1, it has been hypothesized that both genes might be involved in two opposite strategies of host-bacterium interaction. On the one hand, bap positive cells would trigger biofilm formation without causing any special injury in the host. Alternatively, toxin harbouring cells would lead to an acute infection and cause damage in host tissues, permitting the spreading of the infectious focus to other body sites [6].
6 Transcriptional and functional regulation of Bap
It seems reasonable that the synthesis of a large protein like Bap, which comprises a considerable metabolic effort, has to be strictly regulated and probably coordinated with the production of other elements of the biofilm matrix. In this context, a recent study has shown that the global virulence regulator SarA acts as an activator of bap expression [9]. In a previous study, we and others discovered that disruption of SarA downregulates the transcription of the ica operon and totally impairs PIA/PNAG-dependent biofilm formation [10,11]. SarA can modulate gene transcription through a direct interaction with the target gene promoter or activating the agr regulatory cascade [12]. In the case of the bap gene, gel shift and DNase I footprinting assays led to the conclusion that SarA directly binds to the bap promoter region. Besides, it has been shown that the disruption of agr did not have any effect on bap transcription, indicating that the mechanism through which SarA regulates bap is agr-independent [9].
Apart from transcriptional regulation, Bap functionality is also subject to modulation by the presence of calcium. The existence of four potential calcium binding motifs in the sequence of Bap prompted us to investigate the regulatory role of this metal cation in the Bap-dependent biofilm development. First, we showed that addition of calcium had an inhibitory effect on biofilm formation capacity in a bap-positive strain culture. Curiously, the concentrations of calcium responsible for this behaviour were similar to the concentrations of free calcium found in milk. Mutational studies showed that disruption of two of the four putative EF-hand motifs resulted in a recombinant strain that was capable of forming a biofilm, but whose biofilm was not inhibited by calcium. Based on these results, it was proposed a model in which the Bap conformation and the subsequent biofilm development would change depending on the calcium concentration. At a low calcium concentration (micromolar), Bap would be able to mediate cell aggregation, whereas in the presence of high amounts of calcium (millimolar), the metal binding would cause a Bap conformational change that would render the protein not competent for biofilm formation [7].
7 Relationship between bap and mobile genetic elements
Horizontal gene transfer is one of the principal mechanisms driving the evolution of micro-organisms. In biofilm context, it is widely accepted that the existence of ideal environmental conditions for genetic exchange within microbial communities definitively contributes to the spread of virulence or antibiotic resistance genes. In fact, preliminary work developed by J. Ghigo [13] had evidenced that natural conjugative plasmids induce biofilm development, which favours the infectious transfer of the plasmid. Pathogenicity islands have also been recognized as mobile carriers of virulence-associated gene clusters [14]. The bap gene is carried by a transposon-like element, inserted in a pathogenicity island (SaPIbov2), which is mobile without the presence of a helper phage. An argument against the possibility that the bap gene had spread among different coagulase negative staphylococcal species through SaPIbov2 transfer is that the bap gene is not contained in PAI in coagulase negative staphylococci [3]. With respect to the SaPIbov2 mobility, we have evidenced that SOS induction by subinhibitory concentrations of antibiotics such as fluoroquinolones or β-lactams increases the PAIs transfer frequency among S. aureus, promoting the spreading of the virulence genes contained in the PAI [15,16].
8 Esp
While investigating the role of the Bap protein in Staphylococcus biofilm formation, BlastP searches had revealed the existence of a remarkable structural similarity between the staphylococcal protein and the enterococcal protein Esp. Esp is a high molecular weight surface protein whose frequency is increased among infection-derived Enterococcus faecalis isolates [17].
9 Esp structure
The amino terminal region from Esp, up to residue 743, shares 33% of identity with the region B from Bap. Then, two repeat units constitute the core region. The first one consists of three nearly identical 84 aa A tandem repeats, which do not show significant similarity with any known sequence on the genomic data bases. Contiguously to this region, there is a 69 aa spacer region followed by a series of nearly identical 82 aa C tandem repeats. The C repeat regions of Esp and Bap share 33% sequence identity and show similarity with the repetitive surface alpha and Rib proteins, which have been shown to be a key immunogenic determinant on group-B streptococci [18]. The Esp C-terminal domain contains a membrane-spanning hydrophobic region and the LPXTG motif found in most wall-associated proteins of Gram-positive bacteria [19] (Fig. 2). In contrast to Bap, Esp contains neither the EF-hand motifs for interaction with calcium nor the dimerization domains, suggesting that both proteins perform their functions through different mechanisms.
10 Role of Esp in Enterococcus faecalis biofilm formation
A pioneer study to analyze the relationship between the presence of the Esp protein and the ability to produce biofilm in E. faecalis showed a strong association between both factors [19]. However, in the same study, it was shown that disruption of the esp gene completely impaired biofilm formation capacity in two E. faecalis strains, whereas it did not affect the biofilm formation capacity of a third strong biofilm producer, E. faecalis strain, suggesting the existence of esp-dependent and esp-independent mechanisms for biofilm development in E. faecalis. Later on, independent studies from different laboratories have confirmed the existence of an esp-independent biofilm formation pathway in E. faecalis [20–23]. Another result supporting the role of Esp in biofilm formation is that complementation experiments of esp-negative E. faecalis strains with a plasmid harbouring the esp gene bestowed the capacity to produce biofilm [19,24].
As it had previously happened during the study about Bap, the possibility that the number of A or C repeats could affect Esp functionality was considered. As expected, we did not found any association between the number of A or C repeats and the biofilm formation capacity in the E. faecalis esp-positive isolates [19]. Using an in-frame gene-fusion strategy, a recent report has shown that the minimal functional region contributing to Esp-mediated biofilm enhancement is confined to the non-repeat N-terminal domain. The expression of the chimerical PtrF protein containing the Esp N-terminus in a esp-negative strain leads to the formation of a biofilm indistinguishable from the biofilm developed by the wild type, esp-positive, strain. Moreover, the authors of this study showed that the expression of Esp in a heterologous species (E. faecium), as well as in a heterologous genus (L. lactis), had no effect on the ability of these strains to form biofilms, suggesting the existence of other factors that act synergistically with Esp to enhance biofilm development [25].
11 Role of Esp in Enterococcus faecalis pathogenesis
Given that one third of E. faecalis isolates from patients with bacteraemia and UTIs expresses the Esp protein and that Esp is structurally similar to many surface adhesins involved in binding to host ligands, the possibility that Esp could participate in the interaction of enterococci with uroepithelial tissue seemed very reasonable to Shankar and collaborators. These authors, according to the results obtained in an animal model of urinary tract infection, provided evidence that Esp contributed to colonization and persistence of E. faecalis at this location. However, the presence of the surface protein did not influence histopathological changes associated with acute urinary tract infections [26].
Since it has been demonstrated that Esp is able to confer an increased adhesion capacity, both to abiotic materials [19,27] and biological tissues including Caco-2 cells [28], it is thought to be a key virulence factor of E. faecalis. In view of this point, the possibility that it could facilitate intestinal colonization and translocation to mesenteric lymph nodes was considered by the group of J. Donskey. Measuring the density of wild type and esp-mutant isogenic E. faecalis strains in stool after orogastric administrations, they lead to the conclusion that Esp is not involved in the adhesion to the intestinal epithelium and thus does not participate at this stage of infection [29].
12 Relationship between esp and mobile genetic elements
While examining the genomes of the two isolates from the first vancomycine-resistant E. faecalis obtained from the same patient, strain Shankar et al. [30] showed that cytolysin operon and esp gene were inserted into a 150-kb genomic element that fulfilled the requirements to be considered as a pathogenicity island (PAI). Curiously, esp is located within a 17-kb segment flanked by two IS sequences, which is spontaneously lost in a frequency similar to that observed in modulation of virulence by phase variation. Particularly curious is the fact that, although the esp gene from E. faecium encoding for the Espfm variant protein is also contained in a pathogenicity island, the comparison of nucleotide sequences of the two PAIs have shown that they do not share any common feature, but the esp and araC genes [31]. Hence, once again, the hypothesis of horizontal transfer had to be dismissed. Anyway, these findings suggest that harbouring biofilm-associated genes in such a flexible genetic element like a pathogenicity island could help bacteria in the modulation of their virulent and multicellular responses.
13 BapA
In the effort to identify new factors involved in Salmonella biofilm formation, we searched S. enterica genes encoding for proteins homologous to Bap from S. aureus. The TBLASTN program revealed two ORFs encoding hypothetical proteins with significant similarity with Bap. The first one, stm2689, was annotated as a 11474 bp with a frame shift in position 6969. Through sequencing of the region encompassing this point, we proved that this deletion did not really exist in any of our laboratory strains and therefore stm2689 could be considered as an undisrupted gene encoding for a 386-kDa putative protein. Given that the homology found in silico was functionally confirmed by the effects of increasing and abolishing stm2689 expression on biofilm forming capacity, this gene was renamed as bapA [32].
The second ORF, stm4261, is 16680 bp long and encodes for the biggest protein within the S. typhimurium genome (595 kDa). During the course of our study, stm4261 had been identified as S. typhimurium host-specific colonization factor and renamed as siiE (salmonella intestinal infection) [33]. Differently from what happens with stm2689, neither disruption nor overexpression of stm4261 had any effect on biofilm development in any of the strains and conditions tested. This lack of phenotype could be due to the absence of expression or functionality under the conditions tested. Bearing in mind the case of E. faecalis strains in which the disruption of esp did not lead to a decreased biofilm formation, a hypothetical SiiE deficiency could be counteracted by other proteins, like BapA.
14 BapA structure
Individual pair wise alignments between BapA and Bap showed that the homology was restricted to the C-repeat region of Bap (aa 948–2139) and region B of BapA (aa 159–3003). Region B of BapA contains 29 imperfect tandem repeats of 86–106 amino acids, each of which showed on average a 29% of identity with a C-repeat of Bap (Fig. 2). According to protein domain prediction programs (ProDom Structural Genomics), region B of BapA also exhibits homology with a large repetitive RTX inner membrane family of exoproteins, involved in adhesion and virulence processes. Region C contains three VCBS repeats (aa 3117–3214; aa 3371–3466; aa 3626–3734) (Inter Pro IPR010221) found in several large proteins apparently related with adhesion functions and two three non-interacting α-helices within the 60 terminal amino acids, a structure reminiscent of the C-terminal secretion signal recognized by an ABC transporter involved in type-I protein secretion.
15 Role of BapA in Salmonella biofilm formation
Once we had established the negative effect of bapA disruption on biofilm capacity, the high probability of introducing PCR-associated mutations in such a large sequence drove us to use a plasmid-free approach to perform the complementation with a wild-type functional gene. This strategy, that combines the lambda-red linear DNA recombination method with site-directed insertion of the pBAD promoter upstream the target gene, allowed us to simulate the effect of bapA mutation through glucose-mediated inhibition of gene expression [34]. Surprisingly, the arabinose-mediated bapA induction resulted in the formation of a biofilm pellicle so thick that the tube could even be tilted without spilling the liquid out.
Taking into account the experimental evidence gathered for Staphylococcus biofilm formation, we decided to test the hypothesis that BapA could mediate the development of a biofilm independently from the other two known elements up to now: cellulose and curli fimbriae. Performing complementation studies, we came to the conclusion that neither cellulose nor curli fimbriae deficiency could be compensated by the overproduction of the surface protein BapA. Thereafter, BapA seemed to be essential but not sufficient for biofilm development in S. enteritidis. We also carried out the reciprocal experiment and elucidated that overexpression of curli fimbriae is able to counteract the lack of bapA, leading to a normal pellicle development. Moreover, we proved that, although over expression of cellulose on its own is not sufficient for BapA deficiency counteraction, in the absence of cellulose, the complementation of the bapA-negative strain through curli overproduction does not take place, reinforcing the idea of the indispensable role that this polysaccharide has within the biofilm formation process.
Additionally, these results supported the hypothesis that BapA could play a complementary role to fimbriae in connecting cells, either by strengthening fimbriae-mediated interactions or by allowing the interconnection of bacteria separated by long distances. In fact, we found that, when BapA overexpressing and BapA minus cells were mixed together, the pellicle almost exclusively contained BapA positive bacteria, suggesting that the presence of BapA on the surface of the cell is necessary to mediate bacterial recruitment into the biofilm pellicle. These data are compatible with a model in which BapA might interact with itself through homophilic interactions, acting both as a receptor and as a ligand between two bacterial clusters. Besides, this hypothesis could explain why proteins belonging to different bacterial species that show homology to Bap are able to mediate adherence and biofilm formation in a broad range of hosts and environmental conditions [32].
The presence of a C-terminal potential secretion structure involved in an ABC-transporter mediated secretion, together with the analysis of the contiguous genes that had revealed the presence of three ORFs encoding for proteins exhibiting homology with the components involved in a classical type-I secretion mechanism, set the stage for the study of the effect of these genes on BapA function. As expected, the in-frame deletion of the stm2690–92 region resulted in the absence of pellicle formation, suggesting the necessity for BapA to be secreted in order to participate in the biofilm formation process. According to this result, the genes stm2690, stm2691 and stm2692 were renamed as bapB, bapC and bapD, respectively [32]. The absence of BapA in any of the fractions corresponding to the bapBCD minus strain, together with RT-PCR experiments, proved that bapABCD genes constitute a single operon. Besides, we observed that the band present in the supernatant exhibited a slightly lower molecular weight than the band present in the whole protein extract, leading us to the hypothesis that BapA could be either processed during the secretion process or specifically degraded outside the bacteria [32]. Considering that our efforts to label BapA introducing a flag in the carboxy-terminal extreme have always failed due to the loss of the peptide, the possibility of processing while being transported seems quite reasonable.
16 Transcriptional and functional regulation of BapA
The checkpoint of the genetic cascade responsible for biofilm formation at the air–broth interface of LB is represented by CsgD, a LuxR type regulator that promotes both synthesis of curli fimbriae and cellulose [35]. The finding that BapA was also required for biofilm formation under these conditions leads us to test the hypothesis that its expression could also be regulated by CsgD. Real-time PCR assays confirmed that the expression of bapA is CsgD dependent and therefore coordinated with the other two major components of the Salmonella matrix. Although the mode by which CsgD activates the target genes remains unknown [36], putative conserved CsgD-binding consensus sequences located upstream of the putative -35 elements have been described for csgBAC and adrA genes [37]. In the case of the adrA promoter, this sequence has an almost perfect inverted repeat. Interestingly, a slightly different sequence to one of the inverted repeats of the adrA promoter is present upstream the promoter region of bapA, suggesting that CsgD might directly affect the bapA gene transcription. Alternatively, CsgD could indirectly stimulate bapA transcription via the activation of an intermediate regulatory gene. The hypothetical model for biofilm formation through CsgD regulation is shown in Fig. 3.
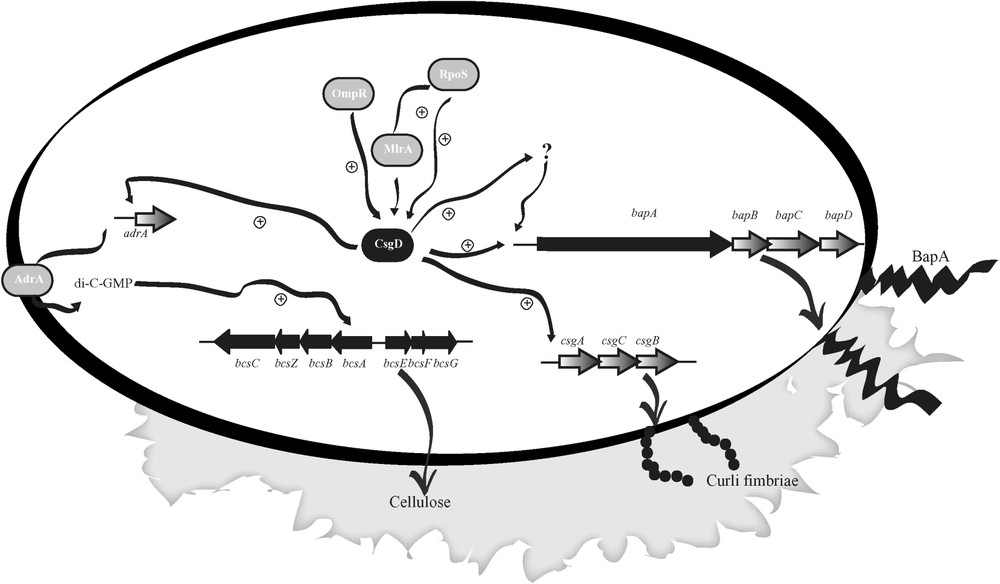
A genetic model for biofilm formation in Salmonella enteritidis. CsgD, a key regulator of biofilm formation on LB, modulate and coordinate the synthesis of fimbriae, cellulose and exoprotein BapA.
Apart from transcriptional regulation and emerging as a novel common feature with Bap, we also have some evidences that BapA is functionally modulated by calcium. Although, in contrast to Bap, motif searches did not reveal the presence of any EF-Hand motif, our preliminary results have shown that BapA-dependent biofilm formation is impaired by the addition of millimolar amounts of calcium to LB medium. Curiously, the core repeated region of BapA exhibits homology with the repeats present in RTX toxins widespread among Gram-negative bacteria, many of which require calcium for its cytotoxic activity [38].
17 Role of BapA in virulence
Using survival, co-infection and ligated ileal loops assays in BALB-C mice, it has been shown that BapA deficient strain is attenuated and shows a decreased ability to invade the intestinal epithelium and to access internal organs [32]. A question that remains open is whether the colonization deficiency is due to the absence of BapA protein itself or to the absence of biofilm. A recent study of Weening et al. [39] provided evidence that deletion of csgA, which is essential for biofilm formation, does not reduce virulence or intestinal colonization. Although this result would be in agreement with a direct role of BapA within intestinal epithelial colonization, further studies are required to clarify the role of BapA in virulence.
18 Relationship between bapA and mobile genetic elements
Even though we have not found any evidence for BapA being contained in a pathogenicity island (SPI), the homolog gene (sty2875) in S. typhi is contained in the SPI-9 pathogenicity island. To date, there is no evidence supporting the mobility of SPI-9.
19 Concluding remarks
Despite of all the experimental evidence supporting a key role of biofilm-associated proteins within adhesion and biofilm formation processes, there are still some unanswered questions we are still looking forward to settle. The first unknown aspect is referred to the mechanism of action of these proteins, since it still remains unclear whether they promote biofilm formation through the interaction with the compounds of the matrix or through a homophilic interaction between two Bap proteins. Another intriguing aspect about Bap proteins is their large size mainly due to the presence of a core domain formed by a variable number of tandem repeats. Considering that the shorter versions obtained both in vitro and in vivo are still functional, there must be a good reason that compensates the metabolic effort that the production of such a large protein involves. Finally, the fact that the genes encoding for these proteins are normally located in mobile genetic elements, like transposons and pathogenicity islands, is particularly interesting. At present, it is difficult to speculate on the benefits that the loss of Bap proteins could provide to the bacteria, but it is conceivable that the loss of Bap enables single bacterium to detach from the biofilm an disseminate into novel habitats.


