1 Introduction
Since the era of Louis Pasteur, Robert Koch and Kitasato Shibasaburō, microbiology has been a vibrant and dynamic research domain covering taxonomy, physiology, biochemistry,… Bacteria, and in particular Escherichia coli, became model organisms to understand fundamental biological questions such as gene regulation and protein synthesis. Molecular biology exploded, invading various areas of research. But there came a time in the late seventies when scientific interest in bacteria decreased and many researchers focused on eukaryotes and higher organisms. Bacteria became more of a tool to clone one's favorite gene, many bacteriologists disappeared and many bacteriology labs closed. Yet, many subjects went on, such as studies on secretion or sporulation. In addition, scientists started to use molecular biology tools to address host-pathogen interactions. Several technological advances emerged, such as high through - put genomics, confocal microscopy, etc., which contributed to resurrect several themes of research. It is clear that microbiology has been undergoing a true renaissance as evidenced by breakthroughs that were unimaginable 10 years ago and more and more young scientists are joining the discipline. Since microbiology is such a vast topic, the organizers wished to focus on aspects of the discipline that could not have been covered in a meeting organized ten years ago, hence the name “the New Microbiology”. The conference was partitioned into several sessions, each of which represents a blossoming field of “New Microbiology”. A variety of international speakers were invited representing some of the up and coming contributors to microbiology as well as long-standing pillars of the microbiology community. In total, there were more than 250 attendees to what was an extremely lively and informative conference.
2 Small RNAs and new regulatory mechanisms
Jörg Vogel (University of Würzburg) investigates small regulatory RNAs in bacterial pathogenesis, specifically in Salmonella typhimurium with emphasis on genome-wide small RNA discovery. Using an approach to isolate novel small RNAs based on their association with Hfq, a highly abundant RNA-binding protein critical for virulence and intracellular survival, Vogel was able to characterize large integrated post-transcriptional networks. Although many small RNAs are widely conserved and are used to regulate conserved functions, there are now examples that they are also recruited to regulate pathogen-specific virulence genes. By characterizing the small RNA SgrS in Salmonella he found that SgrS, which normally regulates glucose homeostasis, also targets the horizontally acquired Salmonella effector SopD but not the closely related gene SopD2. Mechanistically, this was due to a difference in one hydrogen bond in the RNA duplex, which highlights the exquisite specificity of this regulation and has broader implications for conservation of post-transcriptional networks between species and how that may affect virulence during horizontal gene transfer. Jörgen Johansson (Umeå University) discussed the enteric pathogen Listeria monocytogenes during its saprophytic lifestyle. This Gram-positive bacterium is able to survive in a wide range of temperatures, varying oxygen and pH concentrations. Interestingly, Johansson et al. noticed a bull's eye pattern on bacterial colonies that were exposed to light/dark oscillations. While there was no difference in bacterial number or viability between the opaque and translucent rings that form the bull's eye pattern, the opaque bacteria had thicker cell walls and produced more extracellular polymeric substances. The ring formation is dependent on a blue-light sensor as well as the σB factor. A transposon mutagenesis screen identified 60 different genes that abrogate ring formation, many of which are involved in the oxidative stress response. Future work will determine how these signaling pathways interrelate and potentially determine the functional relevance on bacterial fitness in the soil or in conditions important for virulence. Rotem Sorek (Weizmann Institute of Science, Department of Molecular Genetics, Israel) highlighted novel families of toxin-antitoxin systems that act as bacterial “immune defense” against bacteriophages. In order to systematically discover novel toxin-antitoxin systems, Sorek took a very elegant and unbiased genome-wide approach by making use of first generation sequencing methods to identify gaps or “dents” in bacterial genome sequences. These “dents” represent genes whose in-frame expression is toxic to the transformed bacteria. Starting with 388 bacterial genomes, Sorek whittled down the list to genes that fit the toxin-antitoxin rule as selected with a specific algorithm and validated the new toxins by cloning under an inducible promoter. He focused on one toxin-antitoxin system from the screen. The antitoxin component of this system is normally degraded by the host-expressed Lon protease, which in turn is inhibited by a T7 phage protein that blocks its assembly into an active protease. The novelty of the screen comes from its unbiased large-scale identification of toxin-antitoxin pairs, which resulted in exposure of a hitherto hidden layer of the arms race between bacteria and bacteriophages. Steve Busby (School of Biosciences, University of Birmingham, UK) opened his talk on transcriptional regulation in bacteria by discussing how the field had started, where it had been, and where it was going now. He argued that direct pan-genome experimental approaches are crucial to the New Microbiology, and outlined a new method, DNA sampling, that is complementary to chromatin immunoprecipitation, that can be used to identify all of the proteins associated with any DNA target. It was clear that transcription from most E. coli gene regulatory regions is controlled by combinations of activators and repressors, but in cases where genes encode highly toxic products, very often, only repressors are involved. It was noted that, even with E. coli, the goal of predicting the transcriptome simply from the genome sequence had proven elusive. One explanation for this may be that promoter activity is affected by the folding of bacterial chromosomes into the nucleoid. Hence the activity of any promoter is affected by its chromosomal position, and experiments to illustrate this were presented. At some positions, promoters appear especially sensitive to DNA supercoiling or dense local arrays of protein. Active promoters appear to cluster at the edge of the nucleoid. Ultimately, this work demonstrates the need for a better understanding of the role of chromosomal location in transcription and translation in bacteria.
3 Bacterial communities and signaling
Jean-Marc Ghigo (Institut Pasteur, Paris) reported very interesting aspects of bacterial communication via volatile molecules. These types of interactions, which take place in bacterial communities such as biofilms, are mediated by extremely diverse molecules and allow long-range communication. Data on the action of two such compounds were presented. The first, gaseous ammonia was shown to trigger an increase in the resistance of bacteria to various antibiotics. Uptake of ammonia by E. coli was indeed shown to trigger an intracellular increase of polyamine synthesis, which modifies bacterial membrane permeability, thereby altering antibiotic sensitivity profiles. The second, trimethylamine, was shown to increase antibiotic resistance of bacteria by acting on the local pH, which, again, modifies bacterial membrane permeability. Other volatile molecules produced by bacteria were reported to modify various bacterial phenotypes such as growth rate, colony morphogenesis, biofilm formation and pigment production. They represent an underestimated system of communication, which plays an important role in bacteria-bacteria interactions (intra- or inter-species) and probably, in the case of pathogenic bacteria, between bacteria and the infected host. Regine Hengge (Institut für Biologie, Mikrobiologie, Freie Universität Berlin, Germany) detailed the complex signaling networks involved in the switch from motility to sessility in commensal and pathogenic E. coli. During the stages of bacterial growth, flagella and curli fibers are expressed at separate times. Flagella, typically associated with motility are expressed in the post-exponential phase and repressed as cells transition into stationary phase when curli fibers become expressed. The expression or repression of the respective proteins is tightly regulated by distinct sigma factors competing for RNA polymerase core enzyme. In addition, a secondary signaling molecule, bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) is critical for both curli fiber formation and for reducing flagellar activity. Interestingly, curli fibers, which are essential for the complex morphology of colonies grown for extended time, massively accumulate around the top layer of cells, whereas on the lower layer flagellated cells seem to be more prevalent (Fig. 1). Hengge hypothesized that they play an early structural role in the process and plans to explore if this is indeed the case. Finally, similar cellular structures were formed in one of the strains responsible for the 2011 German E. coli epidemic (EHEC O104:H4). It will be fascinating to see if this plays a role in EHEC virulence. Sigal Ben-Yehuda (the Hebrew University of Jerusalem, Jerusalem) reported data on a recently discovered type of bacterial communication, which involves nanotube bridges between neighbouring bacteria (Fig. 1). Transfer of fluorescent soluble molecules as well as membrane markers between adjacent Bacillus subtilis cells connected by these nanotubes suggest a continuity of cytoplasm and membrane between these bacteria. A combination of fluorescent and electronic microscopy approaches allows a detailed characterization of these structures. The existence of nanotubes emanating from the bacterial surface without contacting any adjacent bacteria was also reported, a finding which was proposed to correspond to structures sampling the environment for neighbouring bacteria or for nutrients. The observation of nanotubes connecting bacteria belonging to different species suggests that these structures may represent an important and underestimated form of bacterial communication as they provide a network for direct exchange of various molecules among cells.
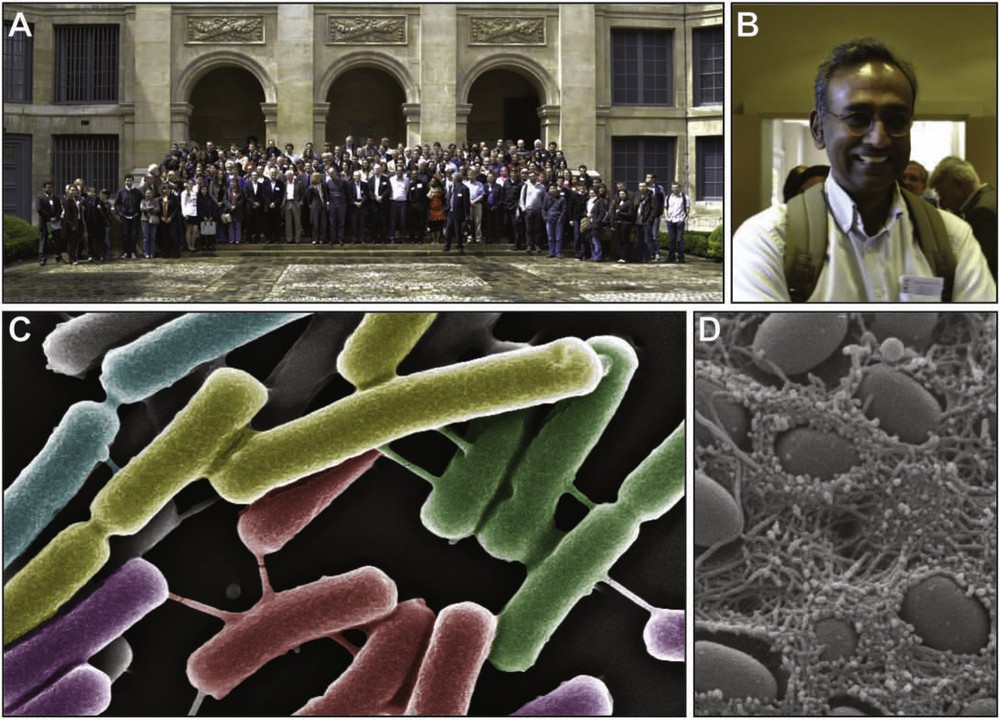
A. The 250 attendees of “The New Microbiology” Symposium in the courtyard of the Institut de France, 14–16 May 2012. B. The Keynote speaker of “The New Microbiology” Symposium and Nobel Laureate, Dr. Venki Ramakrishnan. C. Bacillus subtilis cells grown on solid Luria-Bertani (LB) medium were visualized by using High Resolution Scanning Electron Microscopy (HR-SEM). Nanotubes connecting bacterial cells are visible. Artificial colors were added. Courtesy of Gyanendra Dubey and Sigal Ben-Yehuda. D. Scanning Electron Microscopy (SEM) image of the surface of a 7-day old colony of Escherichia coli K-12 strain W3110. Courtesy of Diego Serra and Regine Hengge (copyright - Diego Serra and Regine Hengge).
4 Microbiota, commensalism and ecology
Eric Pamer (Memorial Sloan-Kettering Cancer Center, New York) presented data on the effects of antibiotic treatment on the intestinal microbiota. Using a combination of both mouse models and clinical data, he convincingly documented that the administration of antibiotics has dramatic effects on both the density and the composition (i.e., the frequency of different bacterial species) of the microbiota. Study of the microbiota during antibiotic treatment indeed revealed a dramatic decrease in the density of intestinal bacteria and although arrest of antibiotic treatment resulted in an increase in bacterial density, the composition of the recovered microbiota remained distinct from that prior to treatment. Antibiotic treatments also alter the host resistance to intestinal colonization by pathogenic bacteria such as Vancomycin Resistant Enterococcus (VRE), a major source of nosocomial infections in humans. Indeed, exogenous administration of VRE to mice treated with antibiotics leads to an efficient colonization of the intestine and to a domination of VRE in the flora, where it reaches frequencies of up to 95%. Once established, VRE remains a dominant component of the microbiota even after discontinuation of antibiotic treatment. In addition, clinical data unveiled that in patients, similar intestinal domination by VRE preceded bloodstream infection. Taken together, these data highlight the potential of high-throughput sequencing of the intestinal microbiota as a means to detect patients at high risk of developing bacterial sepsis. Stanislav Dusko Ehrlich (Inra, Jouy-en-Josas) reported very interesting data on the use of metagenomics to make correlations between genes of the human gut microbiota and human health and diseases, which is the main objective of the European MetaHit consortium. The establishment of an extensive reference catalog of bacterial genes present in the human microbiota was shown to be a key tool to analyse and compare the composition of different human microbiota quantitatively and qualitatively. The comparative analysis of intestinal microbiota from individuals of different countries identified three major clusters called enterotypes, defined by their bacterial composition. These enterotypes, that are not country or continent specific, represent different equilibrated host-microbial symbiotic states, which may respond differently to diet or drug intake. The further identification of a correlation between individuals with a low microbiotal gene content (i.e., with a low diversity of intestinal flora) and certain specific diseases open new research avenues for the development of personalized medicine and/or novel treatments based on altering the composition of the microbiota.
5 Microbial cell biology and single-cell analysis
Kenn Gerdes (Newcastle University, UK) described a mechanism underlying bacterial persistence by toxin-antitoxin systems in E. coli. Persister bacteria are slow-growing variants that appear stochastically in a bacterial population under antibiotic treatments, while remaining genetically identical to antibiotic-sensitive cells. In E. coli K12, mRNA endonucleases (mRNases), which act as toxins and antitoxins, which control the activities of these mRNases, are encoded by several toxin-antitoxin (TA) loci. mRNases are activated in a small fraction of growing cells by the Lon protease that degrades antitoxins. Activation of the mRNases in turn inhibits global cellular translation, and thereby induces dormancy and persistence. In agreement with this, Lon is essential for persistence and deletion of the TA loci reduces persistence. The ability of a single cell to survive ampicillin-induced lysis can be visualized in real-time with a toxin-antitoxin reporter assay. Many pathogenic bacteria known to enter dormant states have a plethora of TA genes. Therefore, the discoveries described here open a door for mechanistic understanding of the persistence phenomenon in various bacterial species. Richard Losick (Harvard University, USA) presented two novel and exciting studies on the mechanisms that govern B. subtilis biofilm assembly and disassembly. The first highlighted crosstalk between tomato plants and Bacillus environmental strains that triggers formation of protective biofilms on plant roots. Plant exudates contain signaling molecules that activate the catch domain of bacterial surface-associated sensor kinases, which in turn phosphorylate the regulator SpoOA, thus activating genes involved in the synthesis of the biofilm extracellular matrix. The second study revealed a double-negative loop that governs stochastic switching between sessile “swimmer” bacteria, which express motility and autolysin genes, and chained “settler” bacteria, which express exopolysaccharide genes. The switch between chained and motile cells is governed by a self-reinforcing loop, in which the R repressor and its inhibitor Slr play a key role. Losick presented beautiful images illustrating the power of microfluidic channels coupled with real-time microscopy to trace single-cell entry and exit from chaining, upon changes in Slr levels, and thus to visualize this regulation loop. Neeraj Dhar from the laboratory of John McKinney (Swiss Federal Institute of Technology in Lausanne, Switzerland) also discussed mechanisms underlying bacterial persistence. He emphasized the notion that persisters, at least in the case of Mycobacterium, are not always slow-growing bacteria with preexisting antibiotic tolerance but rather result from heterogeneity in gene expression at the single-cell level. This concept was addressed by using automated time-lapse fluorescence microscopy of reporter strains of Mycobacterium to follow chromosome replication and cell division upon exposure to the anti-tuberculosis drug isoniazid. Isoniazid is activated by the bacterial enzyme catalase-peroxidase, which was shown to be expressed in apparently random pulses within cell populations. These pulses were demonstrated to correlate with subsequent cell death, consistent with the idea that stochastic fluctuations in gene expression can specify the fate of individual cells. Such mechanisms may allow cell populations to adapt to stressful environments by generating a subset of stress-tolerant individuals. Miroslav Radman (Université R. Descartes, Paris) illustrated the benefit of microbiology to explore novel mechanisms underlying life, death and resurrection, with the example of an exceptionally resistant bacterium, Deinococcus radiodurans. This organism is able to resist extreme desiccation and high doses of UV or ionizing radiation. A key mechanism in this extreme biological robustness is the capacity of this organism to protect its potent DNA repair system from oxidative damage. Deinococcus produces scavengers for oxidation that prevent protein carbonylation. At very high doses of radiation, cell death finally occurs as a consequence of a burst in radiation-induced protein carbonylation and not of DNA damage. A point of no return in cell death is reached when delayed synthesis of oxidized aberrant membrane proteins cause cell membrane depolarization and permeability. Prevention of protein oxidation is thus a tactic for this bacterium to prevent aging and death.
6 Cellular microbiology and manipulation of innate and cellular immunity
Hélène Bierne (Institut Pasteur, Paris) works on the Gram-positive enteric pathogen, L. monocytogenes. Bierne is particularly interested in how Listeria is able to modulate host gene expression through chromatin remodeling during infection of epithelial cells. Listeria accomplishes this goal through diverse means, including the use of a secreted effector, LntA (Listeria nuclear targeted protein A). This protein is present in L. monocytogenes and absent in the closely related non-pathogenic species Listeria innocua. When expressed, LntA translocates to the nucleus of infected epithelial cells and interacts with BAHD1 (Bromo Adjacent Homology Domain Protein 1), a host factor involved in chromatin compaction. BAHD1 forms a complex with KAP1, HP1 and HDAC1/2, among other proteins, and this complex exerts a repressive effect on gene expression. Since overexpression of LntA induced a significant induction of a subset of Interferon Stimulated Genes (ISGs) and, importantly, type III interferon (interferon λ), they hypothesized that BAHD1 may play a role in repressing ISGs during infection. This was indeed the case. An in vivo defect in Listeria colonization of BAHD1 haplodeficient mice further supported the role of this epigenetic regulator in controlling host defense genes. In the future, Bierne et al. plan to further dissect how LntA mediates these effects mechanistically and to continue to characterize the effect of pathogens on chromatin modification. Geoffrey Smith (University of Cambridge, UK) described the astounding capacity of the vaccinia virus to rapidly spread across susceptible cells by a “bounce-like” mechanism. This virus expresses a complex of two proteins on the surface of newly infected cells that, upon contact with superinfecting extracellular virions, stimulated the formation of actin tails which repel these virions. As a result the virions “surf” on the surface of the cell to move to the neighboring uninfected cell. This is an elegant strategy to promote rapid dissemination, by enabling the virus to spread across a lawn of cells without the need to replicate in each cell along the way. Smith also reported identification of the DNA-PK complex as a novel cytosolic sensor for viral DNA. Activation of DNA-PK by double-stranded DNA induces signaling through the STING/TBK1/IRF3 axis, resulting in cytokine production. This discovery points to dual activities of DNA-PK, in both DNA repair in the nucleus and as a pattern recognition receptor to activate innate immune responses in the cytosol. Bruno Lemaitre (École Polytechnique Fédérale de Lausanne, Switzerland) highlighted the power of the Drosophila melanogaster model to dissect and elucidate the molecular mechanisms that govern responses of the gut mucosa to commensal or pathogenic bacteria. Studies in Drosophila enabled the establishment of a key immune role for gut epithelial cells in secreting antimicrobial peptides that destroy pathogens and signaling molecules that activate stems cells, to repair damage associated with the infection. The entomopathogenic bacterium, Pseudomonas entomophila, blocks epithelial renewal in the Drosophila gut via damage caused by the pore-forming toxin monalysin and reactive oxygen species. In their model, epithelial damage induces stress-responsive pathways leading to an arrest of translation that impairs both immune and repair programs in the gut. This indicates that stress-responsive pathways, usually involved in host protection, can become deleterious upon infection. Moreover, this study also shows that inhibition of translation in the gut modulates the innate immune response to intestinal infection: it is useful for host tolerance to commensals or to bacteria with low pathogenic potential, whereas it is deleterious towards bacteria with highly pathogenic potential. Carla Saleh (Institut Pasteur, Paris, France) raised the challenging question of how some animal viruses establish a persistent infection in their insect vectors. She highlighted the necessity of the RNA interference antiviral response in viral persistence, and reported a key role of integration of viral DNA into the host cell genome in boosting the RNAi pathway. More specifically, viral DNA insertions are transcribed and generate small RNAs that activate RNAi antiviral activity, thereby mediating protection against acute infection. In this way, the virus can persistently infect the insect by taking advantage of certain host defenses. Armelle Phalipon (Institut Pasteur, Paris, France) provided new insights into the manipulation of host adaptive immunity by Shigella flexneri. One strategy employed by this enteroinvasive pathogen to dampen the acquired immune defense is to target lymphocytes. Shigella invades activated CD4+T-cells and impairs their migration, both in vitro and in vivo. Moreover, even in the absence of invasion, injection of T3SS effectors upon contact between Shigella and T-cells is sufficient to alter T-cell migration. The key bacterial effector in this inhibition is IpgD, a phosphatidylinositol phosphatase, which reduces the pool of PIP2 at the T-cell membrane, thus promoting dephosphorylation of ERM (ezrin/radixin/moesin) proteins. T lymphocyte polarization and chemotaxis are consequently impaired. These results point to a new mechanism used by a pathogenic bacterium to dampen the adaptive immune response.
7 Genomics, proteomics and synthetic biology
Mark Achtman (University College, Cork) presented new insights in the evolutionary history of Yersinia pestis, the etiological agent of the black plague. Y. pestis is a “genetically monomorphic bacterium”, as it has an extremely low level of genetic diversity. Indeed, the genomes of different strains of this bacterium contain very few polymorphic sites, reflecting unambiguous clonal genealogies. Using genome-wide comparison of different Y. pestis isolates Achtman analysed specific polymorphisms to identify the source and history of different pandemic waves of plague. In particular, the causative strain of the Black Death was proposed to have evolved in or near China and to have spread to areas of other continents on multiple occasions. This type of analysis allows for deeper insight into the etiological agents of historical epidemics without solely relying on historical reports of symptoms. Ruedi Aebersold (Institute of Molecular Systems Biology, ETH Zurich, Switzerland) spoke of a new proteomics approach called Sequential Window Analysis of all THeoretical Mass Spectra (SWATH-MS), which allows for near complete peptide coverage of a bacterial proteome. With this resource in hand, one can subsequently monitor the effects of various stresses or conditions on the entire proteome or focus in on specific peptides, based on their predefined mass to charge ratio, fragment ion patterns and resolution in time. Aebersold went on to validate the approach using Mycobacterium tuberculosis to generate a proteome reference map with nearly 96% peptide coverage. A practical demonstration of the technique compared proteomes from bacteria grown on various carbon sources. Taken together, the experimental possibilities afforded by this technique are vast and may soon become feasible in a variety of different organisms. Philippe Marlière (Isthmus SARL, Paris) presented interesting advances in the field of synthetic biology. This domain of research aims to engineer useful biological systems that do not exist in nature. Xenobiology, one emerging branch of synthetic biology, aims to create orthogonal biological systems based on biochemistry absent in nature. In this vein, data on the modification of DNA with one base replaced by a chemically distinct analog was presented. An automated selection system allowed the creation of an E. coli strain whose DNA contained, in addition to adenine, cytosine and guanine, an artificial base, the thymine analogue 5-chlorouracil. The ultimate goal of this project is to create an organism in which all four nucleotide bases of the DNA are replaced. The growth of such organism would be easily controlled, as it will completely depend on the availability of the incorporated modified bases. Furthermore, such an organism would not compete nor exchange genetic material with “natural” species, thereby allowing the development of a radically distinct class of safe genetically modified organisms without endangering natural habitats or human health.
8 Keynote lecture
Venki Ramakrishnan (Laboratory of Molecular Biology, Cambridge), who shared the Nobel Prize in Chemistry in 2009 for his studies on the structure and function of ribosomes, reported two interesting mechanisms used by bacteria to shut down protein synthesis in response to stress. The first described the structure of the endonuclease RelE, which cleaves mRNA within the ribosome. Ribosomes with such a cleaved mRNA remain stalled due to the absence of a regular stop codon and the resulting recruitment of release factors required for normal termination. Dissociation of ribosomes from these mRNA requires the action of tmRNA (transfer and messenger RNA), a bifunctional molecule acting both as a transfer and a messenger RNA. The tmRNA-encoded open-reading frame is used by stalled ribosomes as a template and restarts the arrested translation. It leads to the addition of a specific tag at the end of the interrupted polypeptide, which plays the role of a degradation signal. Novel structural data on tmRNA association with ribosomes, complexed with its protein partner SmpB, provide a detailed understanding of how these molecules lead to the restart of arrested translation. In particular, SmpB was shown to play the role of both the anticodon loop of tRNA and to portions of mRNA, thereby allowing rescue of translation of cleaved mRNA, without interfering with the translation of regular mRNA.
9 Conclusions
In summary, the conference brought together scientists with a common interest in microbiology and the presentations highlighted the types of hypotheses that are now possible to address thanks to advances in genomics, proteomics, single-cell imaging and microfluidics. A future challenge will be to determine what might distinguish an individual bacterium from its siblings and by extension determining how a bacterium or a small population of bacteria can persist in the face of antibiotics or compete to completely colonize a host. It is likely that work on regulation in single-cell systems combined with next generation sequencing of microbial communities, in the gut for example, will synergize to start to unravel some of these issues. Ultimately, “the New Microbiology” promises numerous fruitful research avenues in the years to come.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
We thank all of the conference presenters and participants for their presentations and for critical reading of this review. “The New Microbiology” was generously sponsored by Danone, La fondation L’Oréal, Institut Merieux, Inserm and the Alfried Krupp von Bohlen und Halbach-Stiftung. We thank Gyanendra Dubey, Sigal Ben-Yehuda, Diego Serra and Regine Hengge for the images shown in the figure. We also thank Eric Postaire and Marie-Thérèse Vicente for their help in organizing the meeting.


