1 Introduction
The order Squamata includes lizards (ca. 4770 species), snakes (ca. 3000 sp.) and amphisbaenians (ca. 170 sp.) [1]. According to recent molecular studies, the closest relatives of snakes are the anguimorphs and/or the iguanians. The presence of toxin secreting oral glands is a shared derived character of this clade (named Toxicofera), demonstrating a single early origin of the venom system in squamates dating from the Jurassic [2,3].
Snakes are divided into two main groups. The fossorial scolecophidians (blindsnakes and threadsnakes, ca. 340 sp.) are small snakes with a limited gape size that feed on small prey (mainly ants and termites) on a frequent basis. The alethinophidians (all other snakes, ca. 2640 sp.) are more ecologically diverse and most species feed on relatively large prey, primarily vertebrates, on an infrequent basis [4,5]. Among Alethinophidia, the caenophidians (advanced snakes, ca. 2470 sp.) widely use venom in addition (or not) to constriction to subdue their prey, while the remaining alethinophidian snakes (ca. 170 sp.), which do not form a single (monophyletic) group, use constriction only (secondarily lost by some fossorial species) [5–10].
Previous studies using DNA sequences to study caenophidian phylogeny have involved relatively few genes: Heise et al. [11] (two mitochondrial genes), Kraus and Brown [12] (one mitochondrial gene); Vidal and Hedges [9] (one nuclear and three mitochondrial genes), Kelly et al. [13] (four mitochondrial genes); Vidal and Hedges [14] (two nuclear genes); Vidal and David [10] (two nuclear genes); and Lawson et al. [15] (one nuclear and one mitochondrial gene). Accordingly, several basal nodes have proven difficult to resolve with statistical significance. For this reason, we have collected additional sequence data and have focused our attention on nuclear protein-coding genes, which have yielded more consistent results in other studies of higher-level squamate phylogeny [2,14,16]. In this study, we address higher-level caenophidian phylogeny with seven nuclear protein-coding genes: oocyte maturation factor (C-mos), recombination activating protein-1 (RAG1), recombination activating protein-2 (RAG2), Orphan G protein-coupled receptor R35 (R35), homeobox A13 (HOXA13), JUN protein (JUN), and amelogenin (AMEL). We direct readers to our earlier studies [8,9] and to Lawson et al. [15] for a more extensive discussion of the taxonomic history of this group.
2 Materials and methods
Tissue samples were obtained from the tissue collections of Nicolas Vidal and S. Blair Hedges (see Vidal and Hedges [5,9] and Vidal et al. [17] for details of the samples used). DNA samples from Pseudoxenodon bambusicola and Calamaria pavimentata were kindly provided by Robin Lawson. Our taxon sampling represents all major caenophidian lineages. DNA extraction was performed using the DNeasy Tissue Kit (Qiagen). The sets of primers used for amplification and sequencing are listed in Appendix A. Both strands of the PCR products were sequenced using the BigDye sequencing kit (Applied Biosystems) in the ABI Prism 3100-Avant Genetic Analyser, or at the Genoscope. The two strands obtained for each sequence were aligned using the BioEdit Sequence Alignment Editor program [18]. Accession numbers of sequence data obtained from GenBank are listed in Appendix B. The 122 new sequences have been deposited in GenBank (Appendix C).
Sequence entry and alignment (25 taxa) were performed manually with the MUST2000 software [19]. Amino acid properties were used, and ambiguous gaps deleted. This resulted in 561 bp for the C-mos gene, 510 bp for the RAG1 gene, 705 bp for the RAG2 gene, 708 bp for the R35 gene, 414 bp for the HOXA13 gene, 372 bp for the JUN gene, and 351 bp for AMEL. In all analyses, remaining gaps were treated as missing data.
We built phylogenies using probabilistic approaches, with Maximum Likelihood (ML) and Bayesian methods of inference. ML analyses were performed with PAUP*4 [20]. Bayesian analyses were performed with MrBayes 3.1 [21,22]. For ML methods, an appropriate model of sequence evolution was inferred using ModelTest [23], both for separate and combined analyses. As we used only protein-coding nuclear genes, and because separate analyses showed no statistically significant topological incongruence, we performed combined analyses, which are considered to be our best estimates of phylogeny. For the concatenated data set (3621 sites), the model selected was the HKY85 + I + G model with a ti/tv ratio of 2.5276, a gamma parameter of 0.9007, a proportion of invariant sites of 0.263, and base frequencies as , , and . Bayesian analyses were run with model parameters estimated as part of the Bayesian analyses, and the best-fit models as inferred by Modeltest. For the ML analyses, we used heuristic searches, with starting trees obtained by random addition with 100 replicates and nearest-neighbour interchange (NNI) branch swapping. For bootstrap ML analyses, we performed 1000 replicates (NJ starting tree with NNI branch swapping). Bayesian analyses were performed by running 2 000 000 generations in four chains, saving the current tree every 100 generations. The last 18 000 trees were used to construct a 50% majority-rule consensus tree.
3 Results and discussion
3.1 Higher-level caenophidian relationships
Our nuclear data set allows us to resolve, with strong support, all major caenophidian splits (Fig. 1). The higher-level topology is virtually identical to the one obtained by Vidal and Hedges [9], but the robustness is significantly increased, which emphasizes the usefulness of nuclear protein-coding genes for resolving deep cladogenetic events.
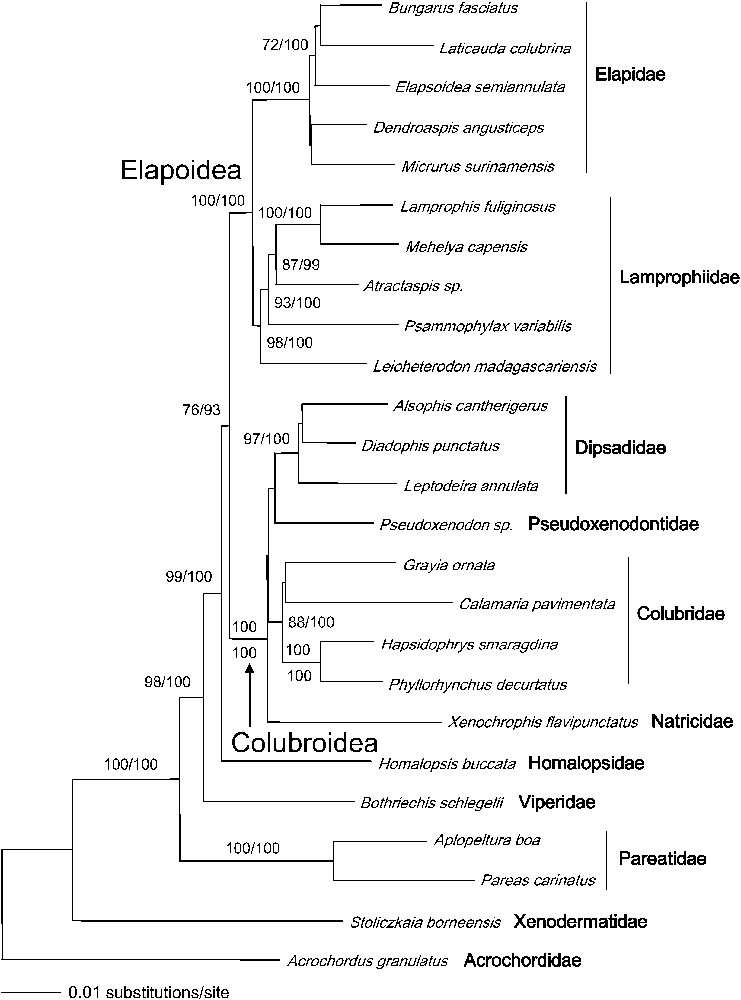
ML tree obtained from the combined dataset (3621 sites, log-likelihood, −15 728.533 97). Values are bootstrap ML values above 70%, followed by Bayesian posterior probabilities above 90%. The ML and Bayesian trees are identical.
Caenophidians have an Asian origin as the following basal lineages, all of Asian range, display a successive branching pattern (basal to derived): acrochordids, xenodermatids, pareatids, viperids (partly Asian), and homalopsids [9]. In several previous molecular studies, xenodermatids were found to be the most basal caenophidian lineage after acrochordids (xenodermatid genera used: Stoliczkaia and Xenodermus) [9,10,14]. An earlier study by Kraus and Brown [12] found a similar position (xenodermatid genus used: Achalinus). Taken as a whole, a basal position for xenodermatids has therefore been obtained for three out of the six extant genera: Achalinus, Stoliczkaia, and Xenodermus. In contrast, using the xenodermatid genus Oxyrhabdium, Lawson et al. [15] obtained a much more derived position, close to elapids and/or the mostly African lamprophiids. As there is no unambiguous morphological character uniting xenodermatids [24], the position of Oxyrhabdium should be further investigated using DNA from another sample in order to confirm the result by Lawson et al. [15]. Because Xenodermus is the type genus of Xenodermatidae, the family name applies to that basal caenophidian lineage including at least two other genera: Achalinus and Stoliczkaia.
Caenophidians can be inferred to have a nocturnal origin, because species belonging to the basal lineages (acrochordids, xenodermatids, pareatids, viperids, and homalopsids), with the exception of some viperids, are primarily nocturnal. All diurnal caenophidians with an active foraging mode and a high metabolic rate are found within the remaining clade, which is divided into two main groups.
The first group comprises the cosmopolitan natricids, the cosmopolitan colubrids (also including the African genus Grayia and the Asian calamariines), the Asian pseudoxenodontids and their closest relatives, the American dipsadids. The second main group comprises elapids and lamprophiids, an African lineage comprising ‘lamprophiines’ (Lamprophis and Mehelya in our study), psammophiines (Psammophylax), Madagascan ‘pseudoxyrhophiines’ (Leioheterodon), and atractaspidines (Atractaspis).
3.2 Taxonomic implications
A conservative classification for the main caenophidian lineages is proposed. Caenophidians devoid of a front-fanged venom system were traditionally lumped into a huge (ca. 1875 sp.) and paraphyletic family named ‘Colubridae’, including several subfamilies. Here, we elevate most of those subfamilies to a familial rank (and restrict the name Colubridae to a monophyletic group) in order to reflect their evolutionary distinctiveness.
- Caenophidia Hoffstetter, 1939
- Superfamily Acrochordoidea Bonaparte, 1831
- Family Acrochordidae Bonaparte, 1831
- Superfamily Xenodermatoidea Gray, 1849
- Family Xenodermatidae Gray, 1849
- Superfamily Viperoidea Oppel, 1811
- Family Viperidae Oppel, 1811
- (Subfamilies: Causinae Cope, 1860, Viperinae Oppel, 1811, Azemiopinae Liem, Marx and Rabb, 1971, and Crotalinae Oppel, 1811)
- Superfamily Pareatoidea Romer, 1956
- Family Pareatidae Romer, 1956
- Superfamily Homalopsoidea Bonaparte, 1845
- Family Homalopsidae Bonaparte, 1845
- Superfamily Elapoidea Boie, 1827
- Family Elapidae Boie, 1827
- (Subfamilies: Elapinae Boie, 1827 and Hydrophiinae Fitzinger, 1843)
- Family Lamprophiidae Fitzinger, 1843
- (Subfamilies: Psammophiinae Bonaparte, 1845, Atractaspidinae Günther, 1858, “Lamprophiinae” Fitzinger, 1843 and “Pseudoxyrhophiinae” Dowling, 1975)
- Superfamily Colubroidea Oppel, 1811
- Family Colubridae Oppel, 1811
- (Subfamilies: Colubrinae Oppel, 1811, Calamariinae Bonaparte, 1840, and Grayiinae, nov. tax.) Grayiinae, nov. taxon, of the family group, with subfamilial rank, with the genus Grayia Günther, 1858 as type genus. Included genus: Grayia
- Family Natricidae Bonaparte, 1840
- Family Pseudoxenodontidae McDowell, 1987
- Family Dipsadidae Bonaparte, 1840
- (Subfamilies: Heterodontinae Bonaparte, 1845 (former North American xenodontines), Dipsadinae Bonaparte, 1840 (former Central American xenodontines) and Xenodontinae Bonaparte, 1845 (former South American xenodontines).
Acknowledgements
We thank those persons and institutions who contributed tissue and DNA samples used in this study: B. Branch, L. Chirio, K. Daoues, I. Das, H. Dowling, J.-C. de Massary, A. Halimi, R. Highton, S. Imbott, U. Kuch, S. Lavoué, R. Lawson, O. Pauwels. This work was funded by the ‘Service de systématique moléculaire, Institut de systématique’ (CNRS FR 1541) to N.V., by grants from the NASA Astrobiology Institute and N.S.F. to S.B.H., and by the ‘Consortium national de recherche en génomique’, Genoscope (France).
Appendix A Primers used
Amplification and sequencing was performed using the following sets of primers:
L39, 5′–CTG–SAR–YTT–TCT–YCA–TCT–GT–3′ [5], HC3, 5′–CAA–ACA–TTA–YRT–TCT–GTG–ATG–A–3′ [5] and G74, 5′–TGA–GCA–TCC–AAA–GTC–TCC–AAT–3′ [25] for the C-mos gene; L2408, 5′–TGCACTGTGACATTGGCAA–3′ [14], H2928, 5′–GACTGCYTG GCATTCATTTT–3′ [14] and H2920, 5′–GCCATTCATTTTYCGAA–3′ [14] for the RAG1 gene; L562, 5′–CCT–RAD–GCC–AGA–TAT–GGY–CAT–AC–3′ [2] and H1306, 5′–GHG–AAY–TCC–TCT–GAR–TCT–TC–3′ [2] for the RAG2 gene; L29, 5′–CTG–AAA–ATK–CAG–AAC–AAA–A–3′ [2], L29B, 5′–CTG–AAA–ATG–CAG–AAC–AAA–AGT–AC–3′ [2], L42, 5′–GAA–CAA–AAG–TAC–WGT–TTC–AAT–3′ [2], L75, 5′–TCT–AAG–TGT–GGA–TGA–TYT–GAT–3′ [2], H786, 5′–TTG–GRA–GCC–ARA–GAA–TGA–CTT–3′ [2], H792, 5′–CAT–CAT–TGG–RAG–CCA–AAG–AA–3′ [2], and H792B, 5′–CAT–CAT–TGG–GAG–CCA–RAG–AAT–GA–3′ [2] for the R35 gene; F2, 5′–ATC–GAG–CCC–ACC–GTC–ATG–TTT–CTC–TAC–GAC–3′ [26], F35, 5′–GTC–ATG–TTY–CTY–TAC–GAC–AAC–AG–3′ [2], F54, 5′–ACA–ACA–GCY–TGG–ARG–AGA–TYA–ACA–A– 3′ [2], R2, 5′–TGG–TAG–AAA–GCA–AAC–TCC–TTG–3′ [26], and R2B, 5′–TGG–TAG–AAA–GCA–AAC–TCC–TTG–G–3′ [2] for the HOXA13 gene; LJUN, 5′–CAG–TTC–YTS–TGC–CCC–AAG–AA–3′ [2], and HJUN, 5′–GAC–TCC–ATG–TCR–ATR–GGG–GA–3′ [2] for the JUN gene; LAM2D, 5′–TAY–CCA–CRK–TAY–DSY–TAT–GAR–CC–3′ [2], LAM2N, 5′–TAT–CCA–CGT–TAT–GGC–TAT–GAA–CC–3′ [2], and HAM, 5′–CAC–TTC–YTC–YTK–CTT–GGT–YT–3′ [2] for AMEL.
Appendix B Sequence data obtained from GenBank
Acrochordidae: Acrochordus granulatus (RAG1 (R): AY487388, C-mos (C): AF544706), Xenodermatidae: Stoliczkaia borneensis (R: AY487398, C: AF544721), Pareatidae: Aplopeltura boa (C: AF544715); Pareas carinatus (C: AF544692), Viperidae: Bothriechis schlegelii (R: AY487374, C: AF544680), Homalopsidae: Homalopsis buccata (C: AF544701), Elapidae: Bungarus fasciatus (R: AY487389, C: AF544732); Dendroaspis angusticeps (R: AY487395, C: AF544735); Elapsoidea semiannulata (R: AY487373, C: AF544678); Laticauda colubrina (R: AY487404, C: AF544702); Micrurus surinamensis (R: AY487411, C: AF544708), Lamprophiidae: Lamprophis fuliginosus (R: AY487378, C: AF544686); Leioheterodon madagascariensis (R: AY487377, C: AF544685); Mehelya capensis (R: AY487379, C: AF544703); Psammophylax variabilis (R: AY487380, C: AF544709); Atractaspis micropholis (C: AF544677), Atractaspis corpulenta (R: DQ993174), Pseudoxenodontidae: Pseudoxenodon karlschmidti (C: AF471102), Dipsadidae: Diadophis punctatus (R: AY487403, C: AF544705); Leptodeira annulata (R: AY487375, C: AF544690); Alsophis cantherigerus (R: AY487376, C: AF544694), Colubridae: Hapsidophrys smaragdina (R: AY487381, C: AF544691); Phyllorhynchus decurtatus (R: AY487385, C: AF544728); Calamaria pavimentata (C: AF471103); Grayia ornata (C: AF544684), Natricidae: Xenochrophis flavipunctatus (C: AF544714).
Appendix C Sequence data produced for this work
Acrochordidae: Acrochordus granulatus (RAG2, R35, JUN, AMEL), Xenodermatidae: Stoliczkaia borneensis (RAG2, R35, HOXA13, JUN, AMEL), Pareatidae: Aplopeltura boa (R35, HOXA13, JUN, AMEL); Pareas carinatus (RAG2, R35, HOXA13, JUN, AMEL), Viperidae: Bothriechis schlegelii (RAG2, R35, HOXA13, AMEL), Homalopsidae: Homalopsis buccata (RAG2, R35, HOXA13, JUN, AMEL), Elapidae: Bungarus fasciatus (RAG2, R35, HOXA13, JUN, AMEL); Dendroaspis angusticeps (RAG2, R35, HOXA13, JUN, AMEL); Elapsoidea semiannulata (RAG2, R35, HOXA13, JUN, AMEL); Laticauda colubrina (RAG2, R35, HOXA13, JUN, AMEL); Micrurus surinamensis (RAG2, R35, HOXA13, JUN, AMEL), Lamprophiidae: Lamprophis fuliginosus (RAG2, R35, HOXA13, JUN, AMEL); Leioheterodon madagascariensis (RAG2, R35, HOXA13, AMEL); Mehelya capensis (RAG2, R35, HOXA13, JUN, AMEL); Psammophylax variabilis (RAG2, R35, HOXA13, JUN, AMEL); Atractaspis micropholis (RAG2, R35, JUN, AMEL), Atractaspis corpulenta (HOXA13), Pseudoxenodontidae: Pseudoxenodon bambusicola (RAG1, RAG2, R35, HOXA13, JUN, AMEL), Dipsadidae: Diadophis punctatus (RAG2, R35, HOXA13, JUN); Leptodeira annulata (RAG2, R35, HOXA13, JUN, AMEL); Alsophis cantherigerus (RAG2, R35, HOXA13, JUN), Colubridae: Hapsidophrys smaragdina (RAG2, R35, HOXA13, JUN, AMEL); Phyllorhynchus decurtatus (RAG2, R35, HOXA13, JUN, AMEL); Calamaria pavimentata (RAG1, RAG2, R35, HOXA13, JUN, AMEL); Grayia ornata (RAG1, RAG2, R35, HOXA13, JUN, AMEL), Natricidae: Xenochrophis flavipunctatus (RAG2, R35, HOXA13, JUN, AMEL).


