1 Introduction
The biological functions of DNA adenine methylation have been widely investigated in Salmonella typhimurium. Mutants lacking DNA adenine methylase show increased spontaneous mutations, moderated SOS induction, enhancement of duplication segregation, inviability of dam recA and dam recB mutants, suppression of the inviability of the dam recA and dam recB combinations by mutations that eliminate mismatch repair [1]. S. typhimunum dam mutants do not show increased UV sensitivity, suggesting that methyldirected mismatch repair does not play a role in repairing UV-induced DNA damage. S. typhimunum dam recJ mutants are viable, suggesting that the Salmonella RecJ function does not help repair DNA strand breaks formed in the absence of Dam methylation [1]. Plasmid-encoded fimbriae (Pef) expressed by S. typhimurium mediate adhesion to mouse intestinal epithelium, whose production is an example of methylation-dependent gene regulation [2]. Finally, DNA adenine methyltransferase modulates Salmonella virulence in many species, like nematodes and mice [3].
The -methyladenine (6mA) sites are present in the homologous replication origins of five enterobacterial species such as E. coli, S. typhimurium, etc. [4], suggesting a conserved function for 6mA in bacterial replication. A positive role for dam methylation in oriC replication is supported by the experimental finding that in vitro DNA synthesis is reduced with unmethylated oriC templates such as pOC42 [1,5,6]. The greatly reduced transformation frequencies of S. typhimurium strains lacking the Dam methylase (dam−) by oriC plasmids pOC42 has also been interpreted as an evidence of a stimulatory role of methylation in oriC replication [5,6]. In addition, plasmid pBR322 [7] transforms dam− strains at reduced frequencies [5,6], although the effect is not as pronounced as with oriC plasmids pOC42.
The poor transformation of dam− strains by methylated plasmids, either carrying the oriC origin or not, suggests that the absence of dam methylation can inhibit replication. These results indicate that this origin is not able to function adequately without methylation. The inability of dam− strains to methylate unmethylated molecules could explain their reduced transformation efficiencies. Transformation inhibition of dam− strains is shown to be completely reversed when unmethylated plasmids are used and when hemimethylated plasmids accumulate after one round of replication of fully methylated plasmids in dam− strains. These findings suggest that hemimethylation may play a key role in controlling replication in vivo, providing a mechanism for preventing reinitiation on newly formed daughter molecules.
2 Experimental procedures
2.1 Bacterial strains and plasmids
Table 1 lists plasmids and the genotypes of all strains used in this study. All of the Salmonella typhimurium strains originated from Spain.
2.2 Bacterial culture and transformation
The S. typhimurium strains used in this study are listed in Table 1. Bacteria were grown routinely at 37 °C in an 8 g l−1 fresh sterile liquid medium (Pronadisa, Hispanlab), and aerated by shaking for one night. When necessary, liquid medium was supplemented with ampicillin and tetracycline. Media were solidified by the addition of agar (15 g l−1). Transformation by the calcium chloride method routinely led to 40 ng of plasmid DNA per 200 μl of competent cells [8,9]. Each transformation result in Table 2 is the average of at least two experiments performed with at least two independent isolates of every plasmid type. Competent cells were grown in a 2-fold 8 g l−1 sterile liquid medium (Pronadisa, Hispanlab), and transformants were selected on ampicillin and/or tetracycline plates. Plating for antibiotic resistance was performed at the following antibiotic concentrations: ampicillin at 50 mg l−1 and tetracycline at 15 mg l−1 [10].
Results of transformation by the methylated and unmethylated forms of different plasmids
| Strain | Number of transformants with pBR322 when | |
| methylated | unmethylated | |
| SL1344 (WT) | ||
| SV1610 (dam−) | ||
| Strain | Number of transformants with pUC18 when | |
| methylated | unmethylated | |
| SL1344 (WT) | ||
| SV1610 (dam−) | ||
| Strain | Number of transformants with pOC42 when | |
| methylated | unmethylated | |
| SL1344 (WT) | ||
| SV1610 (dam−) |
2.3 DNA manipulations
Plasmid DNA was purified by the alkaline minipreparation procedure [11]. The DNA concentration was determined by ultraviolet spectrophotometry or by comparison with known standards after gel electrophoresis. The presence or absence of dam methylation was routinely assayed by digestion with DpnI. Unmethylated and hemimethylated DNA were found to completely resist DpnI digestion, as observed on ethidium bromide-stained gels. Digest was done in a 10-fold concentrated REACT® 4 tampon (invitrogen): [10 mM Tris-HCl (pH 7.4); 400 mM NaCl, 0.1 mM EDTA; 1 mM DTT, 200 μg ml−1 BSA; 50% (v/v) glycerol and 0.1% (w/v) TRITON® X-100], and results were analyzed by 0.8% agarose gel electrophoresis.
3 Results
3.1 Transformation with methylated and unmethylated plasmids
Three different plasmids were compared for their ability to transform various S. typhimurium strains: plasmid pBR322, plasmid pUC18, and the 6kb plasmid pOC42, which contains the 2kb PstI oriC fragment cloned into the PstI site of pBR322 [12–14]. The results of transformation by the methylated and unmethylated forms of these plasmids for each strain are presented in Table 2. The frequency of transformation (Tf) was determined in dam+ (SL1344) and dam− (SV1610) S. typhimurium strains. The results are presented as Tf ratios (methylated versus unmethylated transforming DNA) in Table 3.
Relative transformation frequencies of various S. typhimurium strains by methylated and unmethylated plasmids
| Bacterial strains: S. typhimurium | ||
| Plasmid | dam+ | dam− |
| pBR322 | 2.38 | 8.86×10−5 |
| pUC18 | 1.24 | 1.80×10−3 |
| pOC42 | 0.92 | 8.8×10−2 |
Table 2 shows that these three plasmids transform dam+ S. typhimurium equally well, whether methylated or not (approximately 10−5 CFU for each plasmid: values 1 to 6). However, the dam− strains are not transformed nearly as well by the methylated forms of the three plasmids (values: 2′, 4′ and 6′) compared to their unmethylated counterparts (values: 1′, 3′ and 5′). Table 3 illustrates that the Tf ratios in dam− bacteria are inferior to those in dam+ bacteria. The values are approximately 10−5 for pBR322, 10−3 for pUC18 and 10−2 for pOC42. Different plasmids were isolated from transformants of each strain and found to be methylated for dam+ and unmethylated or hemimethylated for dam−, by digestion with restriction enzyme (see Experimental Procedures). Because SL1344 and SV1610 are isogenic except for their dam loci, the difference in their transformation frequencies must be solely due to the dam gene. The absolute level transformation of SL1344 and SV1610 are nearly identical for unmethylated plasmids, except pOC42. This plasmid contains the replication origin of the Escherichia coli chromosome (oriC), and it is characterized by an elevated number of GATC sequences that are methylated at position N6 of adenine by the Dam methylase [6,15,16]. This observation has led to an interesting suggestion that transient hemimethylation of these sequences after the initiation of DNA synthesis could make the origin inactive for further initiations. These results are in marked contrast to those obtained by Russell and Zinder [17] with plasmid pSC101. This plasmid transforms dam+ and dam− E. coli at approximately the same frequency, whether methylated or not. Therefore, the decreased transformation of dam− strains by methylated plasmids is not a general property of all replicons. Because pSC101 has a total of 27 dam sites [18], whereas pOC42 has 33 (22 in pBR322 plasmid and 11 in oriC fragment), it must be the particular characteristics of certain dam sites, and not merely their number that affects transformation (at least some of the dam sites responsible for the poor transformation of dam− strains must therefore lie in the 2kb oriC fragment of pOC42).
3.2 Hemimethylated plasmids accumulate in dam− S. typhimurium
The poor transformation of dam− strains by pBR322, pUC18 and especially by the pOC42 plasmid is a property of fully methylated DNA, but not of unmethylated DNA. In addition, fully methylated or unmethylated DNA can be replicated normally in dam+. In this work, it is shown that dam− strains should be capable of replicating unmethylated plasmids normally, but fully methylated plasmids only once, producing two hemimethylated daughter molecules that do not replicate again, because they are not reactivated by Dam methylase. A second round of replication would produce two hemimethylated and two completely unmethylated daughter molecules. We show that fully methylated plasmids are eventually converted to hemimethylated daughter molecules in dam− S. typhimurium and that most of the DNA is resistant to DpnI and does not replicate when introduced into dam− strains.
Competent cells of dam+ and dam− were transformed with methylated and unmethylated plasmids, and DNA was extracted and analyzed by digestion with restriction enzymes.
Fig. 1 shows the result of such an experiment performed with pBR322. After transformation, plasmid DNA was isolated from different strains and was digested with DpnI, which leaves both unmethylated and hemimethylated DNA. After restriction endonuclease digestion, DNA fragments were separated in horizontal 0.8% agarose gel.
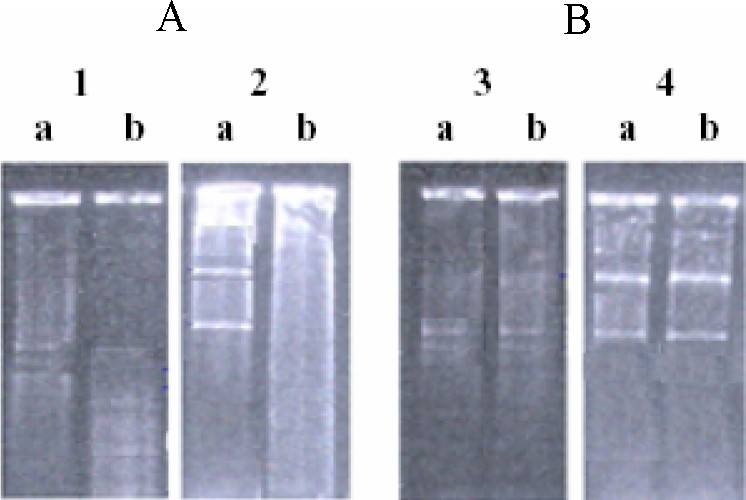
DpnI restriction endonuclease analysis of pBR322 DNA isolated from dam+ (A) and dam− (B) strains: lanes a were the controls without DpnI, lanes b were digested with DpnI.
Sensitivity to digestion by restriction enzyme DpnI indicates that both DNA strands are fully methylated (Fig. 1, lanes 1b and 2b). The intensities of the bands in the two lanes (3b and 4b, Fig. 1) are similar to those in 3a and 4a (Fig. 1), indicating that 4b is completely unmethylated and that 3b shows hemimethylated molecules (made with one methylated strand and one unmethylated strand) that resist digestion by DpnI. Thus, fully methylated pBR322 can replicate one round in dam− bacteria, and the absence of a second round of replication from this methylated plasmid confirms that hemimethylated DNA does not replicate.
4 Discussion
Plasmid pOC42 with the E. coli chromosomal replication origin transform dam− S. typhimurium poorly. This result is very similar to that obtained by Russell and Zinder [17], Smith et al. [5], and Messer et al. [6]. It is shown that the greater number of GATC sequences in pOC42 and the particular characteristics of certain dam sites may explain the greater magnitude of the inhibition of transformation observed compared to pUC18 and pBR322. This inhibition of transformation is completely reversed when the transforming DNA is not methylated (Table 2), so it cannot be due to the absence of methylation in dam− strains. Rather, it must be due to an inhibitory effect of methyl groups present in the transforming DNA. Methylated or unmethylated plasmids transform dam+ strains efficiently (Fig. 2.A.1 and 2), so only using methylated DNA in a dam− strain results in poor transformation. After putting in dam− bacteria, fully methylated plasmids replicate one round and are thereby converted into hemimethylated daughter molecules (Fig. 2.B.2), which are refractory to further replication when compared to their unmethylated counterparts [19]. This result agrees with that of Russell and Zinder [17], who found that hemimethylated plasmid (plasmid heteroduplexes with one methylated and one unmethylated strand, constructed in vitro) transforms GM2199 (dam−) at 1/50 the frequency of GM30 (dam+). Thus, the poor transformation of dam− strains by methylated plasmids can be explained by the accumulation of hemimethylated molecules, which cannot replicate. Therefore, like fully methylated DNA, hemimethylated DNA transforms dam− strains poorly. Dam+ cells can be transformed because methylation of unmethylated DNA creates first hemimethylated and then fully methylated templates for replication (Fig. 2.A.2). Since unmethylated plasmids never become hemimethylated in dam− strains, they can replicate and transform efficiently (Fig. 2.B.1). The finding that unmethylated plasmids transform dam− strains at high frequencies conflicts with the results of Smith et al. [5], who found that unmethylated plasmids transformed dam− E. coli strains at lower efficiencies than their methylated counterparts do. However, as was also observed in this study, Messer et al. [6] found that unmethylated plasmids containing the pMB1 replication origin transform dam− E. coli strains better than methylated plasmids. The reason for these differences is unclear. In conclusion, methylation may play a key role in the regulation of DNA replication in vivo, and generally in eukaryotic and prokaryotic cellular processes [20].
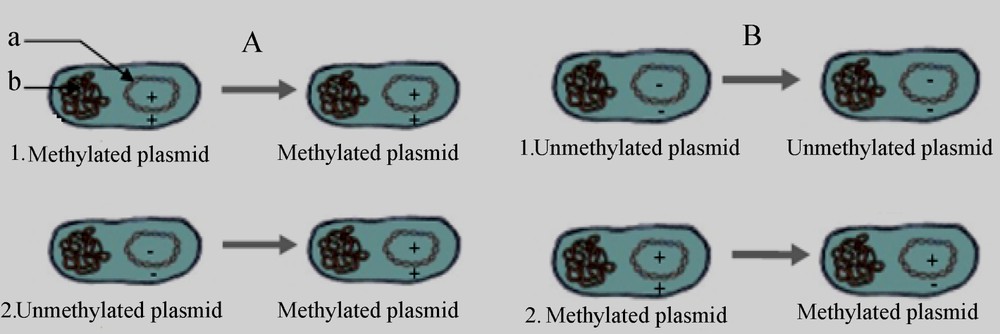
DNA states in different bacterial strains. (a) Transforming plasmid (++: methylated, −−: unmethylated, −+: hemimethylated). (b) Genomic DNA. →: After replication. A: dam+ strains transformed with: fully methylated plasmid (A.1), unmethylated plasmid (A.2). B: dam− strains transformed with: unmethylated plasmid (B.1), fully methylated plasmid (B.2).
Acknowledgements
The authors are grateful to Dr. Francisco Ramos-Morales [21] (Departamento de Genética, Facultad de Biología, Universidad de Sevilla, Spain) for the generous donation of all of the Salmonella typhimurium strains. This work was supported by the Tunisian Ministry of Higher Education, Scientific Research and Technology.


