1 Introduction
The origin of phenotypic novelties is a central question for evolutionary biologists. For understanding how genomic changes can give rise to innovations at more integrative levels, it is mandatory to decipher the mechanisms underlying embryonic development. Evolutionary developmental biology or evo-devo addresses this question by using the knowledge about genes controlling development in model organisms, and comparing developmental processes and their molecular bases in different organisms in a phylogenetic framework [1]. Homology can be assessed and distinguished from homoplasy (parallel or convergent evolution) only by comparing characters in a phylogenetic context. The possibility to investigate the developmental and molecular bases of morphological structures led to redefine homology as a hierarchical concept dependent upon a phylogenetic framework [2], of which consistency at various levels (activator genes, target genes, regulatory networks, developmental processes, morphological structures) can be investigated. Consistency, or lack of consistency, of homology at different scales of complexity may help deciphering the relative part of selection and constraints in the evolution of morphological structures.
In animals, the concept of body plan has contributed to evolutionary analyses of morphological innovations like for instance, analyses of molecular evolution of Hox genes as related with segment formation in Arthropods [3]. It has pinpointed the role of the developmental genetic toolkit in determining the identity, number and pattern of body parts in animals. Most of the toolkit genes consist of transcription factor genes and genes involved in signalling pathways, i.e., genes modifying the expression of other genes. Their functional consequences and further phenotypic impact are highly correlated with their spatial and temporal pattern of expression. In plants, the concept of body plan has been largely ignored until the late 1990s, even if the vascular plants have long been hypothesized to have evolved from a simple body plan that has diversified into the large array of architectures seen today. The study of the molecular and developmental bases of specific traits has long been restricted to a few model species (e.g. Arabidopsis thaliana, Brassicaceae, Antirrhinum majus or snapdragon, Plantaginaceae sensu [4], Oryza sativa or rice, Poaceae), most of which being derived angiosperms. With the increasing influence of evo-devo as an integrative discipline, there is now a trend towards extending the analyses to a broader taxonomic sampling and larger array of traits [5,6].
Seminal evo-devo studies in plants have been dedicated to one of the most remarkable innovation of angiosperms, the flower that may have played a major role in their evolutionary success (more than 235,000 extant species [7]). Considerable work has been devoted to unravelling determinants of the identity of floral pieces, namely sepals, petals, stamens and carpels. Based on mutant studies in the model species An. majus and Ar. thaliana, the ABC model was elaborated [8–10]. The A function determines sepal identity, A + B function, petal identity, B + C function, stamen identity and C function, carpel identity. This model was recently complexified by the discovery of the D function involved in ovule development, and the E function that interacts with the A, B and C functions [11]. Interestingly, all but one of the genes specifying the organ identity belong to a single family of transcription factors, the MADS box genes of MIKC type [12].
Although the genetic determinants of the basic floral ground-plan are now well established [13–15], the molecular bases underlying the large variation in flower architecture (shape, number, colour and synorganization of floral parts) (see Fig. 1) are far from being deciphered. Floral symmetry participates in the attractiveness and beauty of the flowers to the human eye. It is also an integrative architectural trait that account for the astonishing diversity of flower form. Two main types of symmetry are recognized, actinomorphy (polysymmetry, i.e., radial symmetry) and zygomorphy (monosymmetry, i.e., bilateral symmetry). Zygomorphic flowers appear relatively late in the fossil record (late Cretaceous, ) compared to the accepted period for angiosperm origin (early Cretaceous, ) [16–18]. Actinomorphy is considered as the ancestral state for angiosperms, and basal angiosperms mainly have actinomorphic flowers [19]. Zygomorphy evolved several times independently from actinomorphy throughout the angiosperms with multiple reversals towards actinomorphy [20,21]. Changes in floral symmetry may result in changes in efficient pollinator range, which in turn can set up sexual barriers leading to speciation [17]. Accordingly, as a derived architectural trait generally associated with lineage diversification, zygomorphy constitutes a morphological innovation. In fact, some of the most speciose taxa possess zygomorphic flowers [22], such as Fabaceae and Asteraceae in the core eudicots, Orchidaceae in the monocots (see Fig. 1). As a component of the pollination syndrome [23], floral symmetry has probably an adaptive value, and may have coevolved with other traits of the flower. Until now, very few studies have been devoted to this aspect of coevolution of floral traits, although it might help to appraise the relative contributions of adaptation and constraints in the emergence of zygomorphy.

A glimpse at the diversity of floral forms. a: Iris pseudacorus (Iridaceae). b: Centaurea cyanus (Asteraceae). c: Anthyllis montana (Fabaceae). d: Lamprocapnos spectabilis (Fumariaceae). e: Orchis simia (Orchidaceae). f: Passiflora coerulea (Passifloraceae). g: Digitalis purpurea (Plantaginaceae). h: Lobelia tupa (Campanulaceae). i: Canna occidentalis (Cannaceae). Polysymmetric flowers: a, inner flowers of b, f. Monosymmetric flowers: outer flowers of b, c, e, g, h. Disymmetric flower: d. Asymmetric flower: i. Photographs g and h represent flowers and one of their associated visitors. Photographs: Florian Jabbour (a, b, d, f, g, h), Sophie Nadot (c, e), Bruno Lascaux (i).
Floral symmetry is thus worth being considered as a case study for evo-devo. It is a multiform and integrative morphological trait, genetically determined, set up at various stages during flower development. Moreover, mutants for floral symmetry exist in nature or can be obtained in the lab, enabling molecular genetics analyses. In this review, we will give a quick overview of the diversity of floral symmetry in relation with development, before addressing the question of the correlated evolution between symmetry and other floral traits. Then the focus will be put on the molecular bases of zygomorphy in model species that have allowed candidate genes for studies in non-model species to be pinpointed. Accordingly, a large part of this review will be dedicated to a state of the art about the diversity and evolutionary history of these candidate genes and what can be anticipated about their role in floral symmetry. Finally, some of the major questions that have to be addressed in the future to get a better knowledge of the origins and evolution of floral symmetry will be raised.
2 Diversity of floral symmetry
2.1 Diversity of symmetry types in angiosperm flowers
In its current acception, symmetry mainly characterizes the perianth aspect, even though the most elaborate forms also include modifications at the androecium level (see below). Angiosperm flowers are predominantly symmetric or very rarely asymmetric [17] (see Fig. 1i). It was shown that some pollinators (bees, for instance) are only attracted by symmetric flowers [24–26]. A symmetric flower can be polysymmetric, disymmetric (with two perpendicular symmetry planes) or monosymmetric (Fig. 1). Zygomorphic and disymmetric flowers are commonly found in clades characterized by a closed ground-plan (i.e., where floral organ number and arrangement are fixed within and between individuals) as in core eudicots (Lamiales, Fabaceae, Asterales) and monocots (Orchidaceae and Zingiberales). In contrast, actinomorphic flowers are widespread in open ground-plan clades, like basal angiosperms and early diverging eudicots [17,19]. Zygomorphy involves more or less pronounced shape differentiation between organs within a single whorl (or along spirally inserted organs in rare cases). A strong morphological differentiation at the perianth level is often associated with alterations at the androecium level, including stamen reduction (staminodes) or even abortion [27]. It must be noted that floral symmetry is never perfect and small deviations always exist. The minimum rate of deviation is observed in flowers with a high degree of synorganization (e.g. Orchidaceae and Apocynaceae which are respectively zygomorphic and generally actinomorphic) [17].
Asymmetry can be generated by enantiostyly (the style is deviated to the right or the left side of the flower) [28] or reduction, such as in Cannaceae (Fig 1i) or Valerianaceae. In these cases, asymmetry is not irregularity, but rather a highly complicated and ordered phenotype. Unordered simple asymmetric flowers occur in a few basal angiosperms, with chaotic organization where the innermost perianth organs and the stamens are irregularly arranged [17].
2.2 Changes in symmetry during development
An interesting observation deduced from the growing body of studies of flower development is that the symmetry of a mature flower can be different from symmetry at early developmental stages, and that symmetry of the floral primordium may change during development. Accordingly, an actinomorphic flower can present transitory zygomorphic or even asymmetric developmental stages [29]. Zygomorphy is expressed either from organ initiation with no change during the subsequent ontogeny, or later, as a result of differential growth or heterochrony [30], or formation of superstructures due to synorganisations [17]. Both situations occur in the Asteridae sensu [4]. For example, zygomorphy is the product of organ initiation in the subfamily Rhinanthioidae (now included in Orobanchaceae sensu [4]), but it is preceded by an actinomorphic stage during development in the Antirrhinoidae (now included in Plantaginaceae sensu [4]), Bignoniaceae and Lecythidaceae [30].
Symmetry of the floral primordium can also be influenced by the position of the flower in inflorescences, especially during early stages [17]. In contrast, the symmetry of mature flowers can be independent from phyllotaxis, and flowers with either whorled or spiral phyllotaxis can be actinomorphic or zygomorphic (Fig. 2).
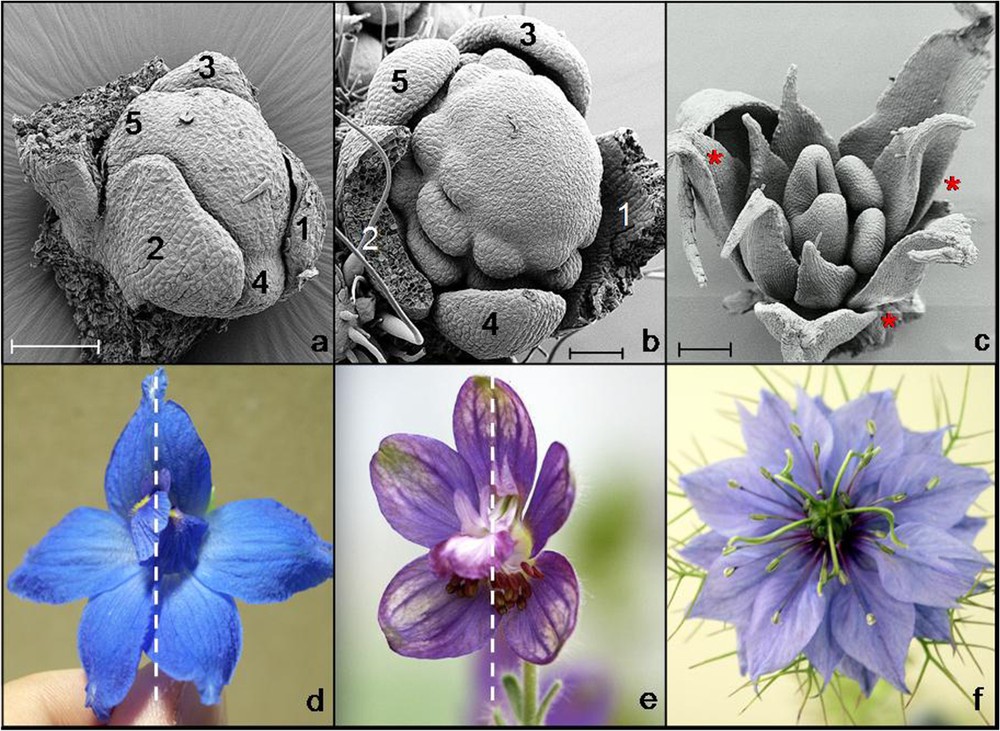
Three examples of buds with spiral phyllotaxis developing into approximately symmetric flowers. (a) and (b) Young buds of Delphinium grandiflorum and Delphinium staphisagria showing the spiral initiation of sepals (scale bars: 100 μm). (c) Late bud of Nigella damascena showing the spiral aestivation of sepals (red stars). (d) and (e) Approximately monosymmetric flowers of D. grandiflorum and D. staphisagria. Dotted lines indicate axes of mirror symmetry. (f) Approximately polysymmetric flower of N. damascena. SEM pictures: Florian Jabbour and Louis Ronse de Craene. Photographs: Florian Jabbour.
2.3 Comparison between the evolution of floral symmetry and the evolution of other floral traits
As previously said, floral symmetry mainly refers to perianth aspect. An interesting question is then whether other architectural traits of the flower, and even further of the inflorescence, can exert a role in the evolution of perianth morphology, either as an initial trigger of adaptive evolution or as steric constraints during development. One possibility to address this question is to examine simultaneously the evolution of several traits of the flower, trying to elucidate the chronology of evolutionary transitions. First results using such an approach were obtained on Ranunculales [31] and on Asteridae [32]. In Ranunculales, zygomorphy appeared three times independently: in the Papaveraceae and the Ranunculaceae, it is associated with the presence of a single spur, while in the Menispermaceae it is limited to the female flowers of some dioecious species. In Asteridae, a phylogenetic comparative analysis revealed that the evolution of perianth symmetry is conditioned by both androecium size and fixed perianth organ number, and that zygomorphy is a prerequisite to the emergence of nectar spurs. The correlation between perianth zygomorphy and oligandry suggests that androecium size might be a developmental or spatial constraint exerted on floral symmetry (Fig. 3), as if a high number of stamens prevented the spatially or temporally heterogeneous development of organs, which characterizes zygomorphic flowers. The results of [31] and [32] revealed different architectural contexts for the evolution of perianth zygomorphy between Ranunculales and Asteridae. In particular, the diversity of evolutionary pathways appears larger in the Ranunculales than in the Asteridae, which may suggest a higher canalization of development in the latter, and possibly different genetic bases in the two clades.
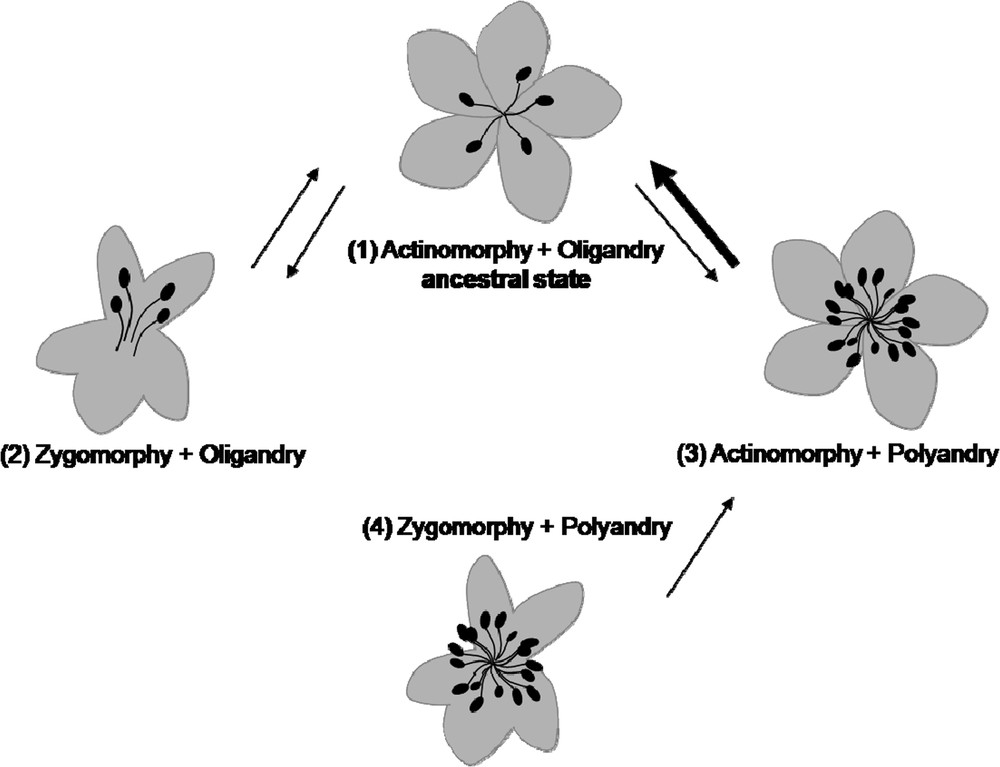
Evolutionary scenario for correlated evolution between perianth symmetry and androecium size in Asteridae (adapted from [32]). Arrows indicate the possible transitions between the associations of traits during the evolutionary history of Asteridae. The bold arrow supports a transition rate significantly superior to the three others. A zygomorphic perianth associated with polyandry was observed very few times in Asteridae.
3 Molecular bases of zygomorphy in model species
3.1 Interplay between four genes in Antirrhinum
Genetic and developmental bases of zygomorphy have been investigated in depth in An. majus, where numerous mutants of flower architecture are available. This species has pentamerous flowers, with five petals fused into a tube ending in two lips, and a dorsal stamen reduced to a staminode. Zygomorphy takes place through dorsoventral asymmetry of the corolla and androecium. The petals are of three types, dorsal, lateral and ventral. Petal type and abortion of the dorsal stamen were found to be controlled by the interplay among four genes belonging to two transcription factor gene families [33,34]. Mutation in the CYCLOIDEA (CYC) gene has the strongest phenotypic effect, with a loss of the dorsal identity and a ventralization of the lateral petals [34]. Together with its paralog DICHOTOMA (DICH), CYC is expressed in the dorsal domain of the young floral meristem, resulting in retarded growth of petals and stamen. At later stages, CYC expression persists throughout the dorsal domain, where it promotes petal lobe growth while it represses stamen development [34]. DICH expression is restricted to the dorsalmost part of the dorsal petals, participating in their internal asymmetry [35]. The dorsal effect of CYC and DICH appears largely mediated by another gene, RADIALIS (RAD). Indeed, RAD has been shown to be activated by CYC in the dorsal domain of floral meristems [36], where it acts antagonistically with DIVARICATA (DIV) that is expressed all over the meristem [33]. DIV is responsible for the ventral identity of petals, which can be observed in the actinomorphic flowers of the cyc-dich double mutant [37,38].
Both DIV and RAD belong to the MYB family. DIV has two MYB-like domains and belongs to so-called R2R3-MYB subfamily [38]. This subfamily appears as the largest in plants, encompassing 97 genes in Arabidopsis for example [39]. RAD has one MYB domain with high similarity with the N-terminal domain of DIV, suggesting that RAD may have evolved from DIV-like genes by C-terminal deletion [33]. It belongs to a recently described subfamily of SANT/MYB proteins [40]. RAD-like genes constitute a small family in both An. majus and Ar. thaliana [40].
CYC and DICH belong to the TCP gene family characterized by a conserved basic helix-loop-helix domain unique to plants [41–44]. TCP proteins were defined by the family founding members, namely TEOSINTE BRANCHED 1 (TB1) in maize, CYCLOIDEA in snapdragon and PCF (Proliferating Cell Factor) in rice [41]. More than 20 TCP genes were found in the completely sequenced genomes of Ar. thaliana and O. sativa [44–48]. Based on the characteristics of the TCP domain, two main classes, I and II, were defined. The resulting proteins were shown to recognize slightly different DNA consensus sequences [49]. Both class I and class II proteins are involved in growth processes. Class I proteins studied up to now function as positive regulators [49–51] whereas class II proteins rather act as negative regulators, even though this role may vary according to the organ or the developmental stage [34,35]. CYC and DICH belong to class II, and share a short conserved domain particularly rich in polar residues, the R domain, which is however not present in all class II genes [41]. In the following, such genes with both domains (TCP and R) will be referred to as CYC-like genes.
3.2 Molecular and genetic studies in Arabidopsis and Lotus
In Arabidopsis thaliana, only five of the 11 class II genes identified, (AtTCP1, AtTCP2, AtTCP12, AtTCP18 and AtTCP24) exhibit an R domain. Phylogenetic analyses [41,42,47] indicate that AtTCP1 could be the closest paralog to CYC. Arabidopsis has an almost actinomorphic flower displaying the tetramerous ground-plan typical of the Brassicaceae (four sepals, four petals, six stamens and two fused carpels). Expression of AtTCP1 was observed only transiently during early stages of flower development. It is expressed asymmetrically, in the dorsal region of early primordia, possibly delaying the emergence of the dorsal sepal primordium [54]. Six RAD-like genes have been characterized in Ar. thaliana. None of them is expressed in the dorsal domain of the flower, in contrast with what was found in An. majus [40]. Transformation experiments indicate that an interaction between CYC and RAD can be obtained in Arabidopsis. However, CYC cannot control the transcription of any of the Arabidopsis RAD-like genes, which suggests that this interaction may have specifically evolved in the Anthirrhinum lineage [36,40].
The genetic bases of zygomorphy have also been investigated more recently into details in the legume species Lotus corniculatus var. japonicus (abbreviated to L. japonicus). Like other Fabaceae species, L. japonicus has strongly zygomorphic flowers with a pentamerous ground-plan. Contrasting with An. majus, the flower of L. japonicus comprises five free petals. The petals are of three morphological types: a large standard formed by the dorsal petal, two lateral wings, and a keel formed by the two joined ventral petals. The androecium is bent, with the dorsal stamens shorter than the ventral ones joined into a tube. It surrounds the pistil and both are enclosed in the keel. The mutant squ1 has flowers bearing an abnormal squared standard, while mutant kew1 has abnormal wings, resembling keel petals [55]. In L. japonicus, four CYC-like genes have been identified. It was shown that the squ1 mutant is affected in the LjCYC2 gene, a close paralog of AtTCP1 and CYC. Further analyses and transformation experiments indicate that LjCYC2 plays a dorsalizing role during flower development comparable to CYC. The double mutant squ1kew1 exhibits fully ventralized flowers, indicating epistatic interactions of LjCYC2 and kew1. The molecular identity of kew1 is unknown at present [55].
4 CYC-like genes as candidates to investigate the origin of floral zygomorphy in eudicots
Analyses in species phylogenetically related to the three previously described model species suggest that CYC-like genes play a general role in the establishment of zygomorphy, at least in core eudicots. For example, hypermethylation of an ortholog of CYC resulting in gene silencing was found to be responsible for the peloric mutant of Linaria vulgaris initially described by Linnaeus [56]. In Mohavea confertiflora, a close relative of An. majus with almost actinomorphic flowers, a change in the territory of expression of the orthologs of CYC and DICH was observed, in a pattern that is consistent with the hypothesis that these genes play a role in the shift in floral symmetry [57]. In Cadia purpurea, a legume species with actinomorphic flowers, an extension of the expression domain of the CYC-like gene LEGCYC1 was observed compared to its zygomorphic close relative Lupinus nanus, consistent with dorsalization of the whole flower [58]. Iberis, one of the few Brassicaceae genera with zygomorphic flowers, displays two ventral petals three to four times larger than the dorsal ones [59]. The ortholog of TCP1 in I. amara (IaTCP1) was found to be expressed symmetrically in young floral meristems, but as development proceeds, the expression becomes stronger in dorsal than in ventral petals, the difference reaching a peak of ∼90-fold after anthesis. Analysis of a peloric mutant with dorsal petals as large as ventral ones indicates a reduced expression of IaTCP1 as compared to wild-type in the corolla, consistent with the hypothesis that IaTCP1 may act to restrain dorsal petal growth in the wild type [59].
CYC-like genes were chosen as candidates to investigate the origin of zygomorphy in the Ranunculales that are considered as the sister group to all other eudicots. Studies have been focused so far on the Papaveraceae [60,61], one of the basalmost families in Ranunculales, which constitutes a morphologically diverse clade in which the genus Pteridophyllum is sister to two subfamilies, Fumarioideae and Papaveroideae. The dimeric ground plan of the flower and especially the perianth with two sepals and two whorls of opposite decussate petals probably offered a favourable context for the development of disymmetric adult flowers through morphological differentiation of the two petal whorls. This was achieved in Fumarioideae, whereas Papaveroideae all have actinomorphic flowers. In addition, in Fumarioideae, disymmetry further evolved into monosymmetry by reduction of one spur. Two paralogous lineages of CYC-like genes were identified in Papaveraceae, named PAPACYL1 and PAPACYL2. They originated through a duplication probably predating the divergence between Papaveroideae and Fumarioideae. Gene expression studies of PAPACYL1 and PAPACYL2 in vegetative and floral organs showed that these two paralogs are expressed during inflorescence and flower development, mainly at organ junctions, and suggested a correlation between PAPACYL expression patterns and flower symmetry [61].
5 Evolutionary history and functional diversification of CYC-like genes
Elucidating the phylogenetic relationships of the CYC-like genes characterized in the eudicots in the context of the evolutionary history of the TCP gene family may help to shed light on the evolutionary forces shaping their diversity and evolution, and to put forward hypotheses about their ancestral function(s) and pattern(s) of expression in relation with floral symmetry.
5.1 Evolutionary history of TCP genes: a story of duplications
Among land plants, sequences belonging to TCP genes were found in mosses (Physcomitrella patens), ferns, gymnosperms [43] and angiosperms. Interestingly, homologous sequences could be amplified with degenerate oligonucleotides in Chara and Cosmidium, two green algae, indicating that the duplication that gave rise to class I and class II genes most probably occurred within Zygnematophyta, before the emergence of land plants [43]. Series of specific duplications appear in land plants, suggesting that this gene family may belong to the toolkit that enabled colonisation of the terrestrial environment [62].
Concerning the class II genes, three main clades comprising monocot and eudicot sequences were tentatively identified (CYC-like or ‘ECE’, CIN-like typified by the CINCINNATA gene of An. majus [63] and a third poorly supported clade), suggesting that at least three class II TCP genes were present in the common ancestor of monocots and eudicots [61]. A complex history of taxon specific duplications and gene losses further took place in each clade [43,48,61,64]. The R domain is present in most sequences of the CYC-like clade, while completely absent from the CIN-like clade. It is very likely homoplastic, since wherever the root of the phylogenetic tree is placed, four events of acquisition/loss are required to account for its distribution.
Focusing on CYC-like sequences in core eudicots, Howarth and Donoghue [65] found three major gene lineages CYC1, CYC2 and CYC3. All sequences were characterized by a short motif of amino-acids in the intermediary domain between the TCP and R domains, leading the authors to propose the denomination of ‘ECE’ clade for all these sequences. Phylogenetic analyses including several monocot sequences as well as basal eudicot sequences (PAPACYL1 and PAPACYL2) are consistent with the hypothesis that at least two of these lineages (CYC2 and CYC3) originated by a duplication at the base of the core eudicots. This duplication may have been genome-wide, since duplications just predating core eudicot emergence were also noticed in several gene families involved in floral development [65]. Enlarged taxonomic sampling is needed to elucidate the evolutionary relationships of core eudicot sequences from the lineage CYC1 with early diverging eudicot and monocot sequences, in order to clarify the history of duplications in the ‘ECE’ clade. It is interesting to note that the AtTCP18 gene that belongs to CYC1 lineage is thought to be an ortholog of the monocot TB1 gene, that played a major role in maize domestication [66]. Up to date in core eudicots, no duplication was recorded in clade CYC1, in contrast to clades CYC2 (containing CYC, DICH, AtTCP1 and LjCyc2) and CYC3, which exhibit numerous more or less recent taxon specific duplications [67] (Fig. 4). In the CYC2 clade, which is hitherto the most widely studied, history of duplications has been elucidated in some cases. Accordingly, the duplication that gave rise to CYC and DICH took place in the ancestor of the tribe Antirrhineae within Plantaginaceae [68], enabling subsequent evolution of the two paralogs by subfunctionalisation [69]. In Legumes, three paralogous lineages (LEGCYC1A and 1B – the latter one comprising the LjCyc2 gene, LEGCYC2) were observed, that probably originated through two successive duplications that took place at the base of the Faboideae, or even before [70,71]. Interestingly in the genus Lupinus, molecular evolution studies suggest that positive selection has been operating in LEGCYC1B lineage in relation to a shift in floral morphology among species [72]. Several independent duplications also occurred within the Asteridae [73]. In Gesneriaceae, a duplication predating the divergence between the subfamilies Cyrtandroideae and Gesnerioideae was found (GCYC1 and GCYC2), and a supplementary duplication took place in the Cyrtandroideae Saintpaulia + Streptocarpus clade [74,75]. In the Dipsacales, the CYC2 and CYC3 paralogs (DipsCYC2 and DipsCYC3) were duplicated in the Caprifoliaceae that comprise mainly zygomorphic species, but not in the actinomorphic sister family Adoxaceae [65,67]. Fig. 4 shows the taxonomic sampling used for CYC-like gene characterisation among eudicots, and the minimum number of paralogs reported in each of the three lineages CYC1, CY2, CYC3, and points to studies that have examined gene expression in relation with floral symmetry (tree adapted from [76]).
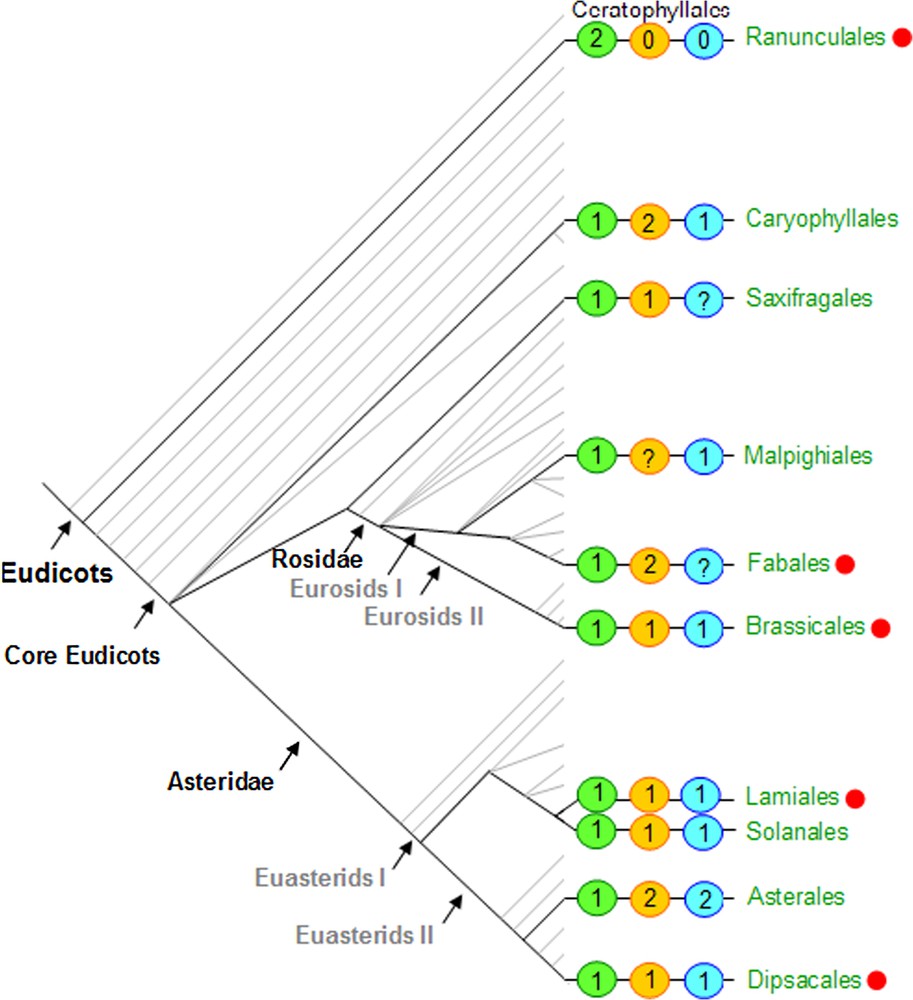
Eudicot tree adapted from the Angiosperm Phylogeny Website [76]. Orders in green include the species in which members of the ECE clade, as described in [65], were found. Orders marked with a red dot are those in which the genetic bases of floral symmetry have been investigated mainly using expression studies (see Table 1). The numbers in the green, orange and blue circles respectively refer to the minimum number of CYC paralogs identified in each of the three lineages CYC1, CYC2 and CYC3, for each species in the order considered.
5.2 Ancestral pattern of expression, function and recruitment for zygomorphy of CYC-like genes
In core eudicots, zygomorphy is considered to have evolved independently in Rosidae sensu [4] and Asteridae from an actinomorphic ancestral state. The results in An. majus and L. japonicus where two CYC2 paralogs are responsible for dorsoventral asymmetry raised an interesting point. In both taxa, did the gene ancestral to the CYC2 lineage have an ancestral function in flower asymmetry? Or were paralogs independently recruited for this trait? More generally, what can be the ancestral function of CYC-like genes, and how did it evolve following the successive duplications, in particular in the three lineages CYC1, CYC2 and CYC3?
The downstream networks of CYC and LjCyc2 are at least partially different, since the RAD gene that belongs to this regulatory network in An. majus, is apparently not involved in the Rosidae examined so far [55]. This appears rather in favor of an independent recruitement of these two paralogs for zygomorphy. Nevertheless, a common pattern of asymmetric expression in lateral organs may exist for CYC2 genes [54]. In An. majus, wild-type inflorescences lack a terminal flower and have zygomorphic axillary flowers. The centroradialis (cen) mutant produces an inflorescence terminated by an actinomorphic flower which resembles the axillary flowers of cyc mutant [77,78]. In this terminal flower, CYC is not active at early developmental stages, but a later transient asymmetric expression is observed in the dorsal regions of the sepals, like in the wild type axillary flowers [79]. In addition to flowers, CYC is expressed asymmetrically in shoots. An asymmetric expression of AtTCP1 was also observed in elongating shoots of Ar. thaliana. In flowers, an asymmetric expression is reported for most if not all CYC2 genes studied (CYC, DICH, Lonicera morrowii DipsCYC2B, LjCyc2, AtTCP1 – see above). An interesting exception appears to be the early expression of the IaTCP1 gene, which could be an apomorphy of I. amara [59].
Only a few data are available concerning the expression and function of genes in CYC3, the CYC2 sister clade. In Lonicera morrowii, a paralog of DipsCYC3 is expressed in flower buds [65]. In Ar. thaliana, AtTCP12 (BRC2 in [80]) prevents axillary bud outgrowth redundantly with AtTCP18, and is also expressed in flowers and siliques [80]. There is no report of asymmetric expression in any of these two cases.
Expression data of genes belonging to the CYC1 lineage in core eudicots, or to related genes in Papaveraceae (PAPACYL) and Poaceae (TB1), indicate an expression in axillary positions and/or at junctions between organs. The TB1 gene and the Arabidopsis AtTCP18 gene (BRC1 in [80]) play a major role in branching. Both genes are expressed in axillary meristems in an apparently radially symmetric pattern, resulting in inhibition of secondary branch growth [52,80]. The TB1 gene in maize also plays a role in sex determination, and a correlation has been found between gene expression and organ (spikelet and especially stamen) abortion [52]. The PAPACYL genes are expressed at organ junctions during flower and inflorescence development. Their particular asymmetric expression observed in the transverse petals in Capnoides sempervirens during development [61] should be examined in other species in order to corroborate this apparent synapomorphy of zygomorphic Fumarioideae. Since the lineage CYC1 and related genes are ancestral to both CYC2 and CYC3, it is tempting to speculate that the asymmetric dorsoventral pattern of expression observed in CYC2 has evolved as an apomorphy of this gene lineage after the CYC2–CYC3 duplication. A proposed ancestral function is growth inhibition or retardation, based on data of several CYC1 genes (see above) and other class II TCP genes [53,63]. Then, sustained asymmetric expression in flowers would be sufficient to promote zygomorphy.
Even though the increasing body of analyses dedicated to the genetic bases of floral symmetry using CYC-like genes as candidates has provided interesting results, our understanding about the origin of this trait during angiosperm evolution is still at its beginning. In particular, no genetic and molecular studies on monocot species, where zygomorphy evolved independently in large clades such as Orchidaceae and Zingiberales, have yet been published. The complexity of class II TCP genes is largely unknown in these clades, the only data in monocots coming from the Poaceae [81]. Moreover, the dorsoventralisation of the flower in An. majus has been shown to rely on the interplay between four transcription factor genes, of which the conservation has not yet been evaluated among the Asteridae. The LjCyc2 interacting partners in Rosidae are still unknown. Widening the sampled taxa and deciphering the gene networks involved during development of zygomorphic flowers are promising perspectives. Some peloric forms appear sporadically in basal eudicots, monocots and basal angiosperms and molecular techniques are now available such as VIGS (Virus Induced Gene Silencing) that enables down-regulation of developmental genes [82–84], which might give the opportunity to test for the role of candidate genes in shaping floral symmetry in various taxa. Other aspects of floral symmetry have to be taken into account in order to progress in the understanding of the evolutionary forces promoting the emergence of this trait among angiosperms. Studying the patterns of symmetry changes during flower development in a phylogenetic context would be of great interest to detect structural constraints and evaluate stability or canalization of development as related to more or less recent transitions towards a zygomorphic phenotype. It would enable to describe the various ways to switch from actinomorphy to zygomorphy, and pinpoint the steps leading to reversals towards ancestral phenotypes. The potential role of “superstructures” in the flower (synorganization of parts) or in the inflorescences should also be challenged. Analyses of relationships between floral symmetry, breeding systems and types of pollinators would help to gain a better view of the role of adaptation in the emergence of particular types of symmetry. Conducting integrative studies that combine molecular genetics, developmental analyses without passing over the ecological aspects is indubitably the key to success in grasping the evolutionary history of floral symmetry.
Acknowledgements
We are grateful to Louis Ronse de Craene for giving FJ the opportunity to conduct SEM studies at the Royal Botanic Garden, Edinburgh. We thank an anonymous reviewer for his constructive comments on the first version of the manuscript. A part of this research (the SEM studies) received support from the SYNTHESYS Project http://www.synthesys.info/ which is financed by European Community Research Infrastructure Action under the FP6 “Structuring the European Research Area” Programme. FJ was supported by a fellowship from the Ministere de l'Enseignement Superieur et de la Recherche, France. We also acknowledge financial support from IFR 87 “La Plante et son Environnement”.


